Protein superstructures can be designed using bottom-up approaches. Genetic engineering and supramolecular chemical strategies are two complementary technologies that are widely used for this. However, genetic methods are prone to high failure rates and often yield unintended structures. In addition, standard genetic methods are restricted to a small set of building blocks, i.e., the twenty canonical amino acids.
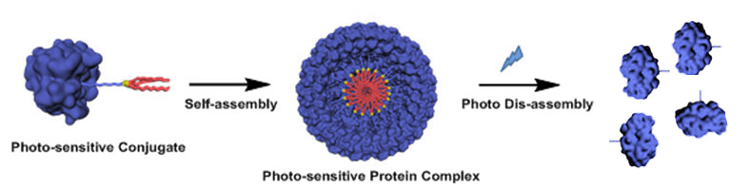
Britto S. Sandanaraj, Indian Institute of Science Education and Research (IISER), Pune, India, and colleagues have developed a supramolecular chemical strategy for the construction of protein complexes. The team used a micelle‐assisted protein‐labeling technology to synthesize libraries of amphiphilic synthetic proteins. These proteins are composed of a hydrophilic globular protein, a flexible hydrophilic linker (both pictured in blue), and a hydrophobic tail (pictured in red). They self‐assemble to form protein complexes through hydrophobic interactions.
The self-assembly of the semi-synthetic amphiphilic proteins was studied using a variety of biophysical techniques, such as size-exclusion chromatography coupled with multi-angle light scattering (MALS), neutron scattering, dynamic light scattering (DLS), zeta potential measurements, and fluorescence and circular dichroism (CD) studies.
The designed protein nanomaterials have a rich structural diversity. Importantly, the team designed the first photo-responsive protein nanoassemblies (pictured), which is important in the area of structural and functional protein nanotechnology. This method complements the existing genetic methods to make protein nanomaterials and offers advantages such as the design of stimuli-responsive protein nanoparticles.
- Rational Design of Supramolecular Dynamic Protein Assemblies by Using a Micelle-Assisted Activity-Based Protein-Labeling Technology,
Britto S. Sandanaraj, Mullapudi Mohan Reddy, Pavankumar Janardhan Bhandari, Sugam Kumar, Vinod K. Aswal,
Chem. Eur. J. 2018, 24, 16085–16096.
https://doi.org/10.1002/chem.201802824