Insights on the mechanism by which natural products and drugs inhibit protein targets can be derived from structural evidence. Understanding structure-activity relationships can lead to more potent drugs. For example, the anti-tuberculosis natural product, griselimycin (GM), isolated from Streptomyces griseus and Streptomyces coelicus and its more active derivative, methylgriselimycin (MGM), only differ by a methyl group on the proline at position 8. These cyclic depsipeptides work by inhibiting the polymerase sliding clamp DnaN of Mycobacterium tuberculosis, the bacterium that causes tuberculosis.
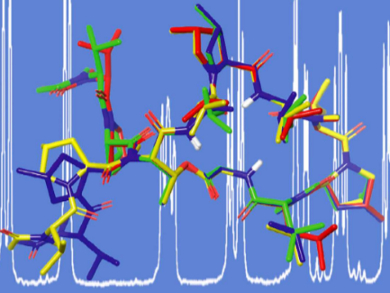
Christine Marie Thiele, Technical University Darmstadt, Germany, and colleagues have used advanced NMR spectroscopy techniques, such as rotating-frame nuclear Overhauser effect (ROE) distant restraints and residual dipolar couplings (RDC), to determine the solution structure of MGM. ROE distant restraints arise from the short-distance coupling between two nuclear spins that are close together in space, whereas residual dipolar couplings arise from the partial alignment of nuclear spins even if they are further away.
The team showed that the ROE and RDC distant restraints allowed for the determination of the backbone conformation of MGM, which was in agreement with the crystal structure of the parent molecule GM bound to DNA polymerase III subunit beta of M. tuberculosis. The RDC measurements detected some flexibility in the structure that is missed out by over-restraining the structure with ROE restraints. Specifically, the RDC measurements were able to show the predominantly down-conformation of the methyl-proline-5 and the more flexible conformation of methyl-proline-8.
This result highlights the sensitivity of RDC to small global structural changes, which can be used in the structure elucidation of other biological molecules. The study underscores the importance of conformational studies on peptides by NMR spectroscopy, especially if no (co-)crystal structure is available.
- Conformational Analysis of an Antibacterial Cyclodepsipeptide Active against Mycobacterium tuberculosis by a Combined ROE and RDC Analysis,
Maic Fredersdorf, Michael Kurz, Armin Bauer, Marc-Olivier Ebert, Carla Rigling, Laurie Lannes, Christina Marie Thiele,
Chem. Eur. J. 2017.
DOI: 10.1002/chem.201605143