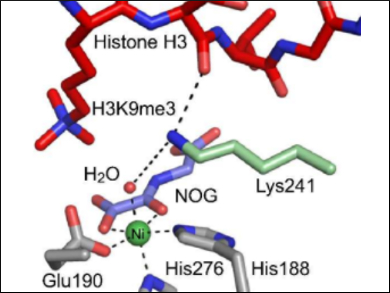
In eukaryotic cells, DNA is compacted through its tight packaging with proteins called histones. The methylation/demethylation of histones, via the amino acid residues lysine and arginine, is an important epigenetic tool to regulate DNA packaging and, therefore, DNA transcription.
Wild-type lysine demethylases (KDMs) catalyze the removal of N-methyl groups by coupling methyl group hydroxylation with oxygen-dependent 2-oxoglutarate decarboxylation. One such KDM, KDM4A, contains a key active-site lysine residue (K241) that is essential for efficient demethylation activity; K241 is proposed to bind oxygen, thus regulating catalysis.
Using mass spectrometry (MS), Richard Hopkinson, University of Leicester, UK, and colleagues have confirmed that K241 is required for efficient demethylation. The team showed that a substitution of K241 with alanine leads to a significantly diminished demethylation activity. However, the decarboxylation of 2-oxoglutarate or the histone binding of KDM4A are not affected, which implies that a regulation of the O2 binding is unlikely.
Overall, the study’s results suggest that lysine K241 regulates coupling of 2-oxoglutarate turnover and histone demethylation. They also indicate that small molecules interacting with K241 could be catalytic inactivators of KDM4A.
- Lysine-241 has a role in coupling 2OG turnover with substrate oxidation during KDM4-catalysed histone demethylation,
Rebecca L Hancock, Martine I Abboud, Tristan J Smart, Emily Flashman, Akane Kawamura, Christopher J Schofield, Richard Hopkinson,
ChemBioChem 2018.
https://doi.org/10.1002/cbic.201800002