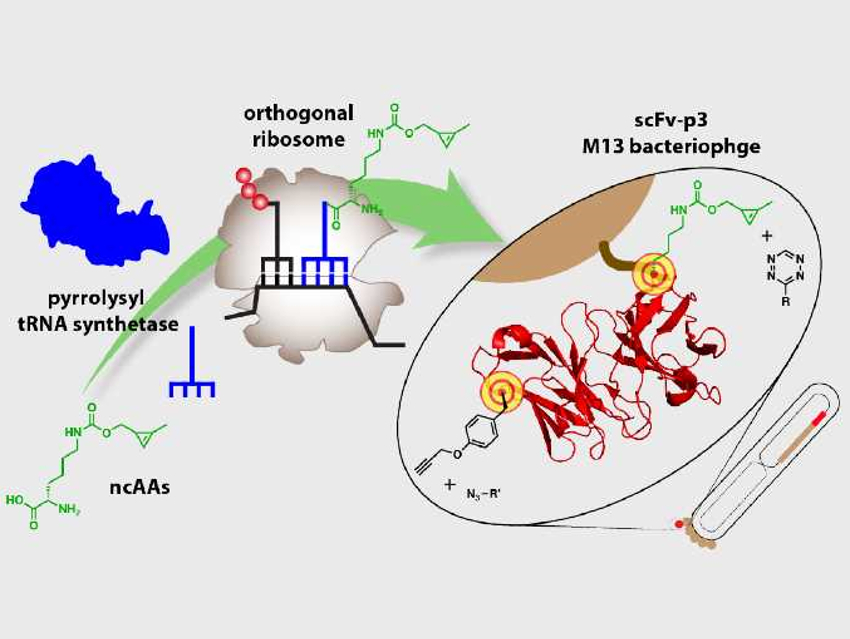
Phage display is a potent technique to produce proteins with new functions. In this method, a protein-encoding gene is inserted into a phage, which displays the encoded protein on the outside. Interactions between the displayed protein and other proteins, peptides, or DNA can then be studied. The approach has been used to produce peptide and antibody therapeutics with a high affinity for certain targets. However, the chemical diversity that can be encoded in phages is mostly limited to the canonical amino acids.
Jason W. Chin and Benjamí Oller-Salvia, Medical Research Council (MRC) Laboratory of Molecular Biology, Cambridge, UK, have developed a system that efficiently incorporates non-canonical amino acids (ncAAs) into phage-displayed proteins. During protein synthesis, messenger RNA (mRNA), a copy of the DNA, provides the sequence of amino acids, and transfer RNA (tRNA) molecules carry the individual amino acids that are connected at a ribosome. This assembly of the amino acids into the final protein is called translation.
The researchers used a translation system that works independently from the natural protein synthesis. They loaded tRNA fragments with the desired ncAAs, which were then translated into the displayed protein. Two different ncAAs with orthogonal functional groups, a cyclopropene-containing lysine derivative (CypK) and p-propargyloxy-L-phenylalanine (PrpF), were both incorporated in the same phage-displayed antibody fragment. Both positions can be tagged with different fluorophores as probes in a one-pot reaction.
According to the researchers, their work expands the number of building blocks and the functional groups that can be encoded on phage-displayed proteins. These advances may contribute to the design of peptide and antibody therapeutics with improved affinity.
- Efficient phage display with multiple distinct non-canonical amino acids via orthogonal ribosome mediated genetic code expansion,
Benjamí Oller-Salvia, Jason W. Chin,
Angew. Chem. Int. Ed. 2019.
https://doi.org/10.1002/anie.201902658