Labeling DNA or RNA using fluorescent nucleotides can allow researchers to monitor the nucleic acid metabolism in living cells. These methods could complement existing metabolic labeling approaches for nucleic acids, which typically require cell fixation and staining. The site‐specific labeling of nucleic acids with fluorescent nucleotide analogues is often performed using solid‐phase synthesis. Reverse transcriptases are enzymes encoded by viruses that catalyze the synthesis of complementary DNA from an RNA template (via an intermediate DNA template) during the replication of the virus. These enzymes could be used for labeling DNA with fluorescent nucleoside analogues instead of solid-state synthesis.
M. Benjamin Turner and Byron W. Purse, San Diego State University, CA, USA, have investigated whether two fluorescent nucleotide analogues called d(tC)TP and d(DEAtC)TP can be used as substitutes for natural deoxycytidine triphosphate (dCTP) in reverse transcription. The two nucleotide analogues contain phenothiazine units as fluorophores; d(DEAtC)TP has an additional 8‐diethylamino (DEA) substituent. The team used the reverse transcriptases from avian myeloblastosis virus (AMV), Moloney murine leukemia virus (M-MLV), and human immunodeficiency virus 1 (HIV-1) to incorporate these nucleotides into nucleic acids. They measured the fluorescent nucleotide insertion kinetics, the elongation efficiency, and the fidelity of the process.
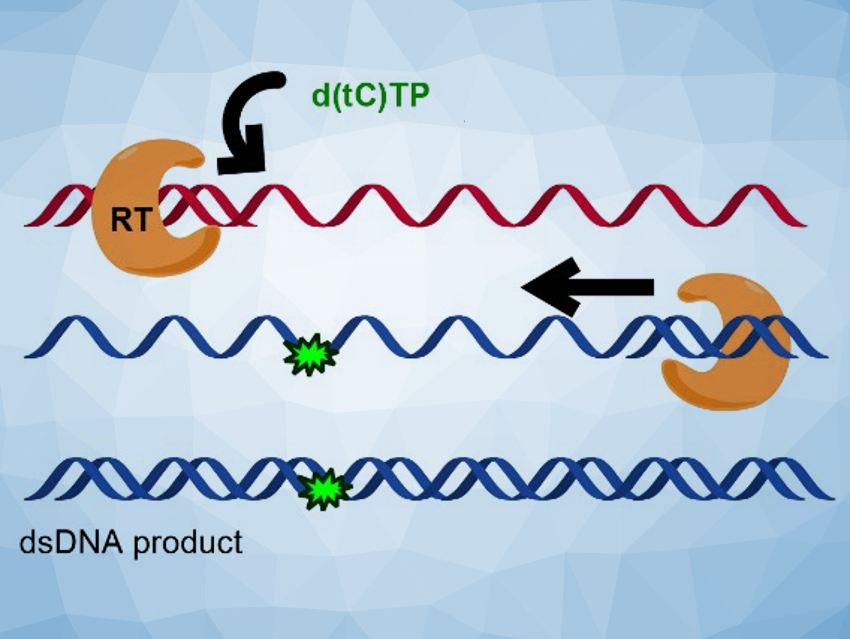
The researchers found that the fluorescent nucleotides are efficiently inserted by all three polymerases, using both RNA and DNA templates. In many cases, the fluorescent cytidine analogues were not significantly distinguished from natural dCTP during the nucleotide insertion. The team performed a full reverse transcription cycle using either d(tC)TP and d(DEAtC)TP and the reverse transcriptase from HIV-1 to convert an ssRNA template (pictured in red at the top) to a labeled dsDNA product (pictured in blue at the bottom). They found that all steps succeeded and full‐length, fluorescently labeled dsDNA could be produced.
- Fluorescent Tricyclic Cytidine Analogues as Substrates for Retroviral Reverse Transcriptases,
M. Benjamin Turner, Byron W. Purse,
ChemPlusChem 2020.
https://doi.org/10.1002/cplu.202000140