Our genome is a chemical text written in our DNA using four letters. According to the blueprints it contains, thousands of different proteins are built from 20 amino acids in the cells of all living beings. However, there are hundreds of natural amino acids. What makes the “magic twenty” so special?
6. Why Only 20 Amino Acids?
The list compiled by Crick and Watson (see chapter 5 in part 1) included only amino acids that are encoded in DNA and are directly assembled into proteins by means of the genetic code in the ribosome. These amino acids are designated as proteinogenic. However, nature presides over a huge repertoire of chemical changes to proteins synthesized in the ribosome. A prime example of this is the oxidation of proline to hydroxyproline in collagen. This reaction is what gives collagen the mechanical properties it needs to act as a support material in our tendons, for example.
There is no scientific reason for the number 20 because individual codons could theoretically encode other amino acids. In fact, after the genetic code was deciphered, two additional amino acids were discovered: #21 selenocysteine and #22 pyrrolysine.
#21: Selenocysteine
In 1986, Stadtman and co-workers [17] discovered that UGA is occasionally not interpreted as the stop codon, instead incorporating selenocysteine (abbreviated Sec). Proteins containing selenocysteine are not common but have been found in all lineages (bacteria, archaea, and eukaryotes). Analysis of the human genome indicates 25 selenoproteins, though not all of these have been identified [18].
The incorporation of selenocysteine happens in a very unusual way. First, the “wrong” amino acid, serine, is bound to the special tRNAsec. While bound, it is then chemically transformed into selenocysteine [19]. This special sec-tRNA[Ser]Sec binds to UAG when a characteristic nucleotide sequence follows the UAG after a specific distance. Only in this case does the tRNA laden with selenocysteine bind to UAG, allowing the selenocysteine to be incorporated into the protein.
#22 Pyrrolysine
Selenocysteine takes a chemical detour to be smuggled into proteins via serine and plays somewhat of an outsider role compared to the “magic twenty”. This is completely not the case for amino acid #22, pyrrolysine. This amino acid (abbreviated Pyr) remained fully unknown until 2002. It was detected in three enzymes in the archaeon Methanosarcina barkeri, which lives at the bottom of freshwater lakes [20,21].
This was not unusual at first glance, because pyrrolysine could have been produced by subsequent chemical modification of an initially “normally” incorporated lysine. However, this was ruled out because pyrrolysine is incorporated with its own tRNApyr [22, 23], like the members of the “magic twenty”, and corresponds to the UAG codon.
Although it so far seems that only Methanosarcina barkeri incorporates pyrrolysine, as of 2002 we have had to speak of the “magic twenty-two”. We will see what chemical structure nature will surprise us with in amino acid #23.
7. Is the Standard Code a “Frozen Accident” or Optimal?
Because the standard genetic code applies to all living things from archaebacteria and bacteria without a nucleus to eukaryotes with a nucleus, it must be very old in evolutionary biology terms, having remained unchanged for many millions of years. Because there is no system evident in this code, Crick called the code a “frozen accident”.
In the 1990s, however, some deviations from the standard code were discovered (see Tab. 4) that indicate that the genetic code must have been changeable in the past.
|
Table 4. Codons with changing meaning. |
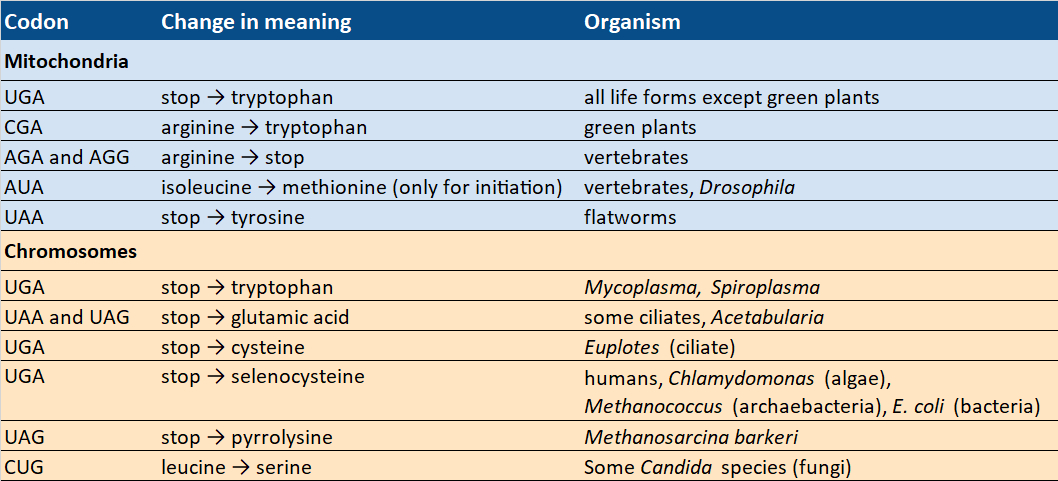 |
At least sixteen species from different kingdoms use modified genetic codes. In fact, we do not need to look any farther than ourselves, because the standard code does not apply everywhere in us. The mitochondria found within our cells—in which most chemical energy is produced, and which are, therefore, known as the powerhouses of our cells—use a variant of the standard code.
Whether the current standard code is a “frozen accident” or the optimized product of evolutionary development was investigated by Stephen Freeland and Laurence Hurst [24]. They began with the following thought: If the current standard genetic code has been optimized through evolution, it must be superior to other conceivable codes. But what does superior mean?
We could begin at the extremes and ask ourselves, how would the best and worst possible genetic codes for evolution look? In the worst case, every mutation, meaning a change in a single base in the DNA, would lead to completely unusable proteins. In the most extreme case, every mutation would lead to nonviable individuals. This type of code would be a dead-end, allowing for no evolution. In the best of all genetic codes, an exchange of bases in the DNA would lead to minimal changes in the functional properties of the proteins, with the associated chance of occasionally finding more effective proteins that are filtered out by natural selection.
We can play through this thought experiment with the AUU codon: If a point mutation changes AUU to AGU, the affected protein would have its nonpolar isoleucine replaced with polar serine. This is a very drastic change with possibly fatal consequences because the hydrophobic character of an amino acid side chain determines the three-dimensional protein structure, and thus, its biological function. In other words, this mutation is likely unfavorable for evolutionary improvement.
However, if AUU is mutated to CUU, then isoleucine is replaced with the similarly nonpolar leucine. This minor change in the protein could have been better tolerated by primeval lifeforms. Such a tiny alteration could be favorable for evolutionary development because tiny changes offer the chance for improvement. Large leaps are usually fatal.
Freeland and Hurst then quantified the hydrophobic character of the side chains in all twenty amino acids and calculated the changes in hydrophobia to be expected in our standard code if each base in all 64 codons were to be changed step by step. The sum of all possible changes is a measure of the total error tolerance or evolutionary capacity of the standard code. Subsequently, the error tolerances for alternative codes were calculated and compared to our standard code. This sounds like a simple computational task, but there are over 1018 alternatives to the standard code, so the calculations are beyond our current technical capabilities. The authors had to settle for random samples of “only” one million alternative codes.
This massive sample gave a surprising result: When reasonable boundary conditions were applied, only one alternative code surpassed our standard code. Such a large error tolerance is indicative of evolutionary selection because life forms with an error-tolerant code not only have a higher chance of survival; they also have the chance to improve their protein reservoir through incremental changes.
Many scientists continue to rave about code without commas, regretting that nature does not conform to it. Perhaps these most recent studies about error tolerance do not yet provide an answer, but they do give an indication that nature does not strive for an abstract, ultimate beauty, but allows for small changes, giving lifeforms the opportunity for evolutionary development. Let us be glad that nature did not listen to either Gamow or Crick, because in Gamow’s diamond code, each change in a base leads to changes in two, or most often even three amino acids, with disastrous consequences for protein function. The code without commas would be even worse because a single altered base most likely goes nowhere: 44 of the 60 triplets are meaningless and would lead to the termination of protein synthesis. It seems we are very well served by our current standard code.
8. Summary
The competition to decipher the genetic code, the transformation of an endless text written with four different symbols into one with twenty symbols, ultimately culminated in the brilliant code without commas developed by Crick, Griffith, and Orgel. For some, it has been disappointing that experiments have demonstrated that nature takes a different, seemingly less elegant chemical route. It has only recently been possible to show that the natural genetic code is far more error-tolerant than alternative codes. Nature is not striving for elegance, efficiency, or superficial beauty; instead, it requires reliability and allows for changes with the chance for improvement.
Acknowledgments
I thank Dr. P. Winchester and Dr. S. Hollman, both of FU Berlin, Germany, for their critical review of the manuscript. I thank A. Peuker, FU Berlin, and T. Roth, HGKZ Zurich, Switzerland, for their help in creating the figures.
References
[17] T. C. Stadtman, Selenocysteine, Ann. Rev. Biochem. 1996, 65, 83–100. https://doi.org/10.1146/annurev.bi.65.070196.000503
[18] A. M. Diamond, On the Road to Selenocysteine, Proc. Natl. Acad. Sci. USA 2004, 101, 13395–13396. https://doi.org/10.1073/pnas.0405357101
[19] R. Wilting, A. Böck, Die Flexibilität des genetischen Kodes, Biol. Unserer Zeit 1996, 26, 369–379. https://doi.org/10.1002/biuz.19960260610
[20] G. Srinivasan et al., Pyrrolysine Encoded by UAG in Archaea: Charging of a UAG-decoding Specialized tRNA, Science 2002, 296, 1459–1462. https://doi.org/10.1126/science.1069588
[21] B. Hao et al., A New UAG-encoded Residue in the Structure of a Methanogen Methyltransferase, Science 2002, 296, 1462–1466. https://doi.org/10.1126/science.1069556
[22] D. G. Longstaff et al., A Natural Genetic Code Expansion Cassette Enables Transmissible Biosynthesis and Genetic Encoding of Pyrrolysine, Proc. Natl. Acad. Sci. USA 2007, 104, 1021–1026. https://doi.org/10.1073/pnas.0610294104
[23] J. M. Kavran et al., Structure of Pyrrolysyl-tRNA Synthetase, an Archaeal Enzyme for Genetic Code Innovation, Proc. Natl. Acad. Sci. USA 2007, 104, 11268–11273. https://doi.org/10.1073/pnas.0704769104
[24] L. D. Hurst, S. J. Freeland, Der raffinierte Code des Lebens, Spektrum Wiss. 2004, 7, 86.
The article has been published in German as:
- Die schönste falsche Theorie der Biochemie. Aus 4 mach’ 20, oder 21, oder 22, oder ….,
Klaus Roth,
Chem. unserer Zeit 2007, 41, 448–458.
https://doi.org/10.1002/ciuz.200700446
and was translated by Caroll Pohl-Ferry.
Deciphering the Genetic Code: The Most Beautiful False Theory in Biochemistry – Part 1
Countless scientists contributed to its clarification with brilliance, ingenuity, intuition, and luck
Deciphering the Genetic Code: The Most Beautiful False Theory in Biochemistry – Part 2
How the experimental verification of the genetic code was accomplished
See similar articles by Klaus Roth published in ChemistryViews