One of the worries about genetically modified organisms is that they will escape the lab and take over the general population. By substituting normal amino acids with artificial or non-canonical amino acids (ncAAs), proteins can be designed which are dependent on the ncAA for activity. This could prevent modified organisms from surviving in the wild, where artificial amino acids are unavailable. This approach has been shown to work in a number of proteins already, but problems remain. The synthetic proteins can easily mutate back to the wild-type protein, or the activity of the protein is greatly decreased because of the substitution, thus making the organism inviable.
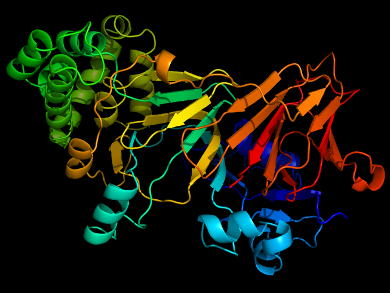
Peter G. Schultz, Scripps Research Institute and California Institute for Biomedical Research, both La Jolla, CA, USA, and colleagues have found that by replacing a histidine residue that is involved in coordination of a zinc cofactor in mannose-6-phosphate isomerase (ManA) with 3-methyl-L-histidine, the enzyme can retain enough activity to function. They showed that E. Coli transformed with the mutant enzyme (H264MeH) was able to grow on mannose as the sole carbon source, meaning the bacteria expressed functional ManA protein.
The team also examined the reversion rate for this mutation and found it to be well below the threshold set for genetically modified organisms by the US National Institutes of Health (NIH). This means that such an organism would not be able to mutate to survive under conditions outside the laboratory. Additionally, the team screened the enzyme for secondary mutations which would enhance the activity of H264MeH. They found one mutation which can restore the activity of H264MeH to a wild-type level. Combined, these two mutations give a fast-growing bacteria that is completely dependent on methyl histidine to survive.
- Functional Replacement of Histidine in Proteins To Generate Noncanonical Amino Acid Dependent Organisms,
Fei Gan, Renhe Liu, Feng Wang, Peter G. Schultz,
J. Am. Chem. Soc. 2018.
https://doi.org/10.1021/jacs.7b13452