DNA-encoded chemical library (DEL) technology is based on the conjugation of molecules to DNA, which then serves as a kind of identification barcode. DELs can have either a single pharmacophore per DNA strand or use a dual pharmacophore model with two complementary strands, each having their own building blocks. The latter enables self-assembly and the formation of combinatorial libraries. These libraries are also called encoded self-assembling chemical (ESAC) libraries. They facilitate the large-scale screening of protein binders (e.g., compounds that bind to kinases, proteases, and phosphatases), which could eventually result in new drug leads.
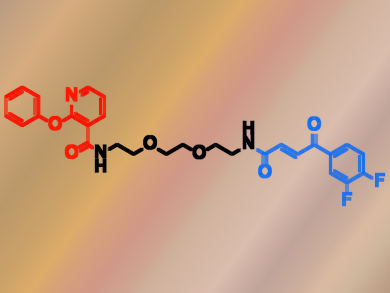
Dario Neri, Swiss Federal Institute of Technology (ETH) Zurich, Martin Mattarella, Philochem AG, Otelfingen, Switzerland, and colleagues have constructed an ESAC library with almost 150,000 members generated through the self-assembly of two sub-libraries. The researchers used the library to screen for covalent JNK1 binders. JNK1 is a kinase that regulates various cellular processes such as proliferation and transcription. The team found a synergistic effect between two building blocks labeled A82 (pictured red) and B272 ( pictured blue) and then verified the activity of these building blocks using mass spectrometry.
The team showed that individual building blocks did not form adducts with JNK1. In contrast, when the two building blocks are covalently linked (A82-L-B272), they form 1:1 adducts with JNK1 with an apparent dissociation constant of 1 µM. The researchers also showed that A82-L-B272 did not interact with other kinases, and thus, is selective against JNK1. In summary, the researchers showed that ESAC libraries could make it easier to discover ligands for proteins of pharmaceutical interest.
- A Specific and Covalent JNK-1 Ligand Selected from an Encoded Self-Assembling Chemical Library,
Gunther Zimmermann, Ulrike Rieder, Davor Bajic, Sara Vanetti, Apirat Chaikuad, Stefan Knapp, Jörg Scheuermann, Martin Mattarella, Dario Neri,
Chem. Eur. J. 2017, 23, 8152–8155.
DOI: 10.1002/chem.201701644