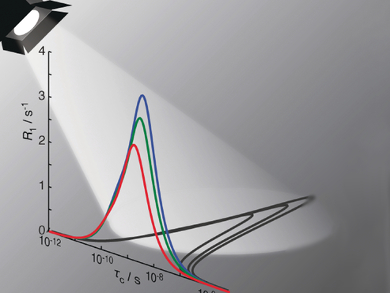
Relaxation in NMR (the return from a non-equilibrium to a normal state) is the result of motion in a sample. Information on the internal motion of molecules can, thus, be obtained by fitting the relaxation data. Typical analyses fit the data to an explicit model that assumes a correlation function between one and three decaying exponential terms, where each term characterizes a motion with a correlation time and amplitude.
Albert A. Smith, Matthias Ernst, and Beat H. Meier, ETH Zurich, Switzerland, have investigated the behavior of such models using theoretical calculations. The team found that when the real motion has more exponential terms than the model, one can successfully fit the data, but significant discrepancies between the real motion and the model arise. In fact, the model fitting is biased towards those correlation times where the relaxation data are most sensitive. This means using explicit models of motion (i.e., a finite number of terms) results in an unreliable characterization of the motion.
To solve this problem, the team introduced a dynamics “detector”, a mathematical expression that can quantify the amount of motion for a range of correlation times, without using an explicit model of motion. These detectors can be used for the bias-free characterization of motion, for example, of proteins.
- Because the Light is Better Here: Correlation-Time Analysis by NMR Spectroscopy,
Albert A. Smith, Matthias Ernst, Beat H. Meier,
Angew. Chem. Int. Ed. 2017.
DOI: 10.1002/anie.201707316