Natural Alternative to Antibiotics
Kyeong Kyu Kim, Sungkyunkwan University School of Medicine, Suwon, Republic of Korea, Seung Seo Lee, University of Southampton, UK, and colleagues have performed a total synthesis of the natural product xanthoangelol B. This compound could be further developed as a drug to inhibit the production of virulence factors in one of the most worrying antibiotic-resistant bacteria, Staphylococcus aureus.
Antibiotic Resistance
Antibiotic pioneer Alexander Fleming was perhaps the first to warn that bacteria would ultimately evolve resistance to the chemical weapons we use against them. Indeed, it was seen very early in the history of penicillin and its descendants that this was the case. Every prescription sees some fraction of the population of microbes that have the necessary defenses to defeat the antibiotics.
The biochemical machinery that the surviving bacteria have devised can spread to wider populations. Whereas a few successful outliers might commonly be overwhelmed in the immunological onslaught against them, there will be genes in the environment that can endow their successors in subsequent infections with the requisite armory to defeat antibiotics wholesale.
We are now rapidly reaching the point where pathogenic microbes, such as S. aureus, Escherichia coli, Clostridium difficile, and many others can easily defeat the frontline of antibiotics in many patients, as well as the second attack formation of antibiotics of last resort. Some pundits will point out that there is little money to be made in pharmaceutical companies developing novel antibiotics. While there is ongoing research in this area, there are no known new potent drugs for mainstream use against infection in the drug company pipelines. Not least because any new antibiotic will follow the same path, with the pathogens rapidly regrouping and evolving the necessary tools to defeat it.
Two-Pronged Attack
What is needed is a new strategy, one that cannot be so easily circumvented by evolution. A strategy that allows a chemical to attack the pathogen in a way that precludes it evolving inhibitors or endogenous agents that defeat it. The researchers were aware that two-component systems could be a strong target in this context. Random mutations that benefit a bacteria when faced with a single agent happen readily, but two random mutations that occur simultaneously to defeat a two-pronged attack are far less likely. This is the basis of fighting infection with two different types of antibiotic at once, but if the genes against both exist in the wild then those random mutations are no longer needed in the light of bacteria sharing and swapping genes.
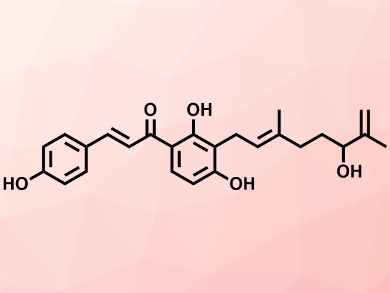
Instead, Mizar and colleagues are focusing on multiple virulence factors in S. aureus as alternative targets for agents that can kill the pathogen. They have screened regulators of these virulence factors in the microbe against natural product inhibitors and homed in on xanthoangelol B (pictured).
This compound is a prenylated chalcone from a type of flowering carrot plant Angelica keiskei, known in Japanese as ashitaba, or tomorrow’s leaf. The team has demonstrated that xanthoangelol B and one of its derivatives, known as PM-56, are both potent inhibitors of the two-component virulence system in S. aureus. They have examined the mode of action of these two compounds and found that they both directly bind to the regulator and inhibit the activity of its associated histidine kinase enzyme. This, the team adds, suggests that the same compounds and other derivatives might act as broad-spectrum inhibitors of histidine kinases.
“Our next step is to derivatize xanthoangelol B and PM-56 by diversity-oriented synthesis to get better activity,” Seung Lee told ChemViews Magazine. “Because we now know what part of the molecule is essential, we are in a position to vary other parts while keeping important structural elements to get a more diverse array of active compounds. In addition, we will test our compounds with histidine kinases and two-component systems from various sources.”
- Total Synthesis of Xanthoangelol B and Its Various Fragments: Toward Inhibition of Virulence Factor Production of Staphylococcus aureus,
Pushpak Mizar, Rekha Arya, Truc Kim, Soyoung Cha, Kyoung-Seok Ryu, Won-sik Yeo, Taeok Bae, Dae Wook Kim, Ki Hun Park, Kyeong Kyu Kim, Seung Seo Lee,
J. Med. Chem. 2018.
https://doi.org/10.1021/acs.jmedchem.8b01012