Copying DNA and the Origin of Life
When a cell divides, it passes on genetic information by producing copies of its DNA. Chemists have also learned to copy DNA. In the journal Angewandte Chemie, a German team has now introduced a new copying technique that uses a single strand of DNA as the “master copy”, like a cell, but does not require enzymes. Unlike earlier methods, it allows for stepwise growth of the chain in both the direction preferred by nature and the opposite direction typical of current DNA synthesis techniques.
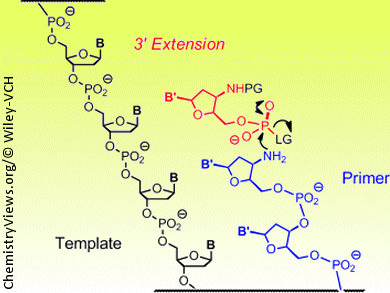
Within a cell, the DNA double strand is separated in segments during the copying process. One of the single strands serves as the “master copy” or template. Polymerase enzymes snap together the corresponding nucleotides stepwise to form the new complementary strand, beginning with a “starting segment” known as a primer. The backbone of a DNA strand is an alternating chain of five-membered sugar rings and phosphate groups. The chain links are formed at the 3’ and 5’ oxygen atoms of the sugars; natural growth occurs in the 3’ direction.
One question relating to the origin of life is: How was nature able to copy DNA or RNA strands before polymerases existed? Since the 1980s, DNA synthesizers have allowed chemists to produce DNA strands, but without a template or primer; the sequence is determined by the order of addition of the reagents. Only the use of protective groups that inhibit uncontrolled reactions and the programmed addition of the reagents ensure that the sequence of bases is correct. This is clearly not how nature does it. But how could template-directed primer extension function purely chemically, with no enzymes?
Chemical Replication with Modified Primer
More recently, different approaches have been used to develop a method called chemical primer extension, which involves the reaction of activated nucleotides with the end of a slightly modified DNA primer. Clemens Richert, Andreas Kaiser, and Sebastian Spies of the University of Stuttgart, Germany, have now developed this method further. They found a protective group that can be removed under gentle conditions so that the DNA duplexes made from the primer and template do not fall apart. This allows the reactivity of the nucleotides and the terminus of the primer to be switched on and off as desired, and the sequence information in the template strand can be read out nucleotide by nucleotide. For this method to work, the template and primer are both attached to tiny spheres. As in an automated synthesizer, the reagents and building blocks can flow over the spheres. The primer is bound to the template through base pairing. A suitable nucleotide from the surrounding solution docks at the next vacant binding site of the template. The nucleotide then binds to the reactive end of the primer through activated phosphate units. The sites that are supposed to react are chemically altered to become more reactive than in natural DNA. The special thing about this method is that the chain extension can be controlled to occur in either the 3’ or the 5’ direction. This is not known to take place in nature.
So far, this process has remained quite slow and is limited to short sequences. Improvement should be possible through optimization of the reaction conditions and better automation.
- Template-Directed Synthesis in 3′- and 5′-Direction with Reversible Termination,
Andreas Kaiser, Sebastian Spies, Tanja Lommel, Clemens Richert,
Angew. Chem. Int. Ed. 2012, 51(33), 8299–8303.
DOI: 10.1002/anie.201203859