The resistance of Gram-negative bacteria to multiple drugs poses a serious threat to global health. One approach to tackling bacterial infections is to focus on inhibiting key bacterial enzymes and their active sites. Gyrase B, an enzyme not present in humans, is a well-validated target for developing antibacterial drugs but currently, no associated inhibitory drugs are on the market.
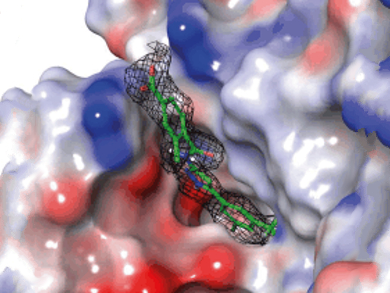
Alvin Hung and colleagues, A*STAR Experimental Therapeutics Centre, Singapore, have applied a combination of biophysical techniques to address this problem. First, a library of 1800 compounds was screened by differential scanning fluorimetry against a truncated version of Escherichia coli gyrase B. Fourteen fragments were identified and their binding was studied by isothermal titration calorimetry, NMR techniques, and X-ray crystallography. Modifications on a low-affinity fragment (KD = 233 µM, IC50 = 628 µM) led to the development of a new class of potent sub-micromolar inhibitors (IC50 = 160 nM) against gyrase B based on the phenyl aminopyrazole scaffold.
The picture shows the co-crystal structure of the pyrazole-based inhibitor in the active site of a gyrase B surrogate. As the urgency for devising new antibiotics grows, these compounds may provide starting points for drug development. Further optimization will be required to generate more potent and cell-permeable bacteria-active compounds.
- Application of Fragment-Based Drug Discovery against DNA Gyrase B,
Guo-Ying Chen, Fui Mee Ng, Yih Wan Tan, Anders Poulsen, Weiguang Seetoh, Grace Lin, CongBao Kang, Siew Wen Then, Nur Huda Ahmad, Ying Lei Wong, Hui Qi Ng, C. S. Brian Chia, Qiu Ying Lau, Jeffrey Hill, Alvin W. Hung, Thomas H. Keller,
ChemPlusChem 2015.
DOI: 10.1002/cplu.201500197
|
This research can be found in a special issue of ChemPlusChem published in honor of Singaporeʼs Golden Jubilee. |