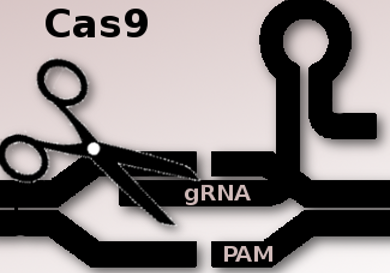
The CRISPR–Cas9 gene editing technique allows researchers to make changes to the genome at specific sites. However, a key limitation is the requirement that a specific DNA sequence, a protospacer adjacent motif (PAM), has to be present at the target site. The most commonly used enzyme Cas9 was isolated from Streptococcus pyogenes bacteria. The required PAM sequence for this SpCas9 is 5′-NGG-3′ where “N” is any nucleobase followed by two guanine (“G”) nucleobases.
David Liu, Harvard University, Cambridge, MA, USA, and colleagues have modified the enzyme to work on more sites in the genome. The team used phage-assisted continuous evolution (PACE) to develop an expanded PAM SpCas9 variant, xCas9. The team tested xCas9 on many sites in the genome. xCas9 can recognize a broad range of PAM sequences including NG, GAA, and GAT, and could target one-quarter of sites in a genome. This is the broadest PAM compatibility reported to date among Cas9s active in mammalian cells. It supports applications in human cells including targeted transcriptional activation, nuclease-mediated gene disruption, and both cytidine and adenine base editing.
Moreover, xCas9 has much greater DNA specificity than SpCas9 and much lower off-target activity. It was less likely to cut at other sites in the genome and produced fewer unwanted effects than the standard laboratory enzyme. According to the researchers, the same approach could be used to modify other variants of the Cas9 enzyme.
- Evolved Cas9 variants with broad PAM compatibility and high DNA specificity,
Johnny H. Hu, Shannon M. Miller, Maarten H. Geurts, Weixin Tang, Liwei Chen, Ning Sun, Christina M. Zeina, Xue Gao, Holly A. Rees, Zhi Lin, David R. Liu
Nature 2018.
https://doi.org/10.1038/nature26155