The discovery of new pharmaceutically active molecules usually involves a purification step of the candidates between synthesis and testing. In activity-directed synthesis (ADS), arrays of reactions with multiple possible outcomes are performed and the crude reaction mixtures are then screened for pharmaceutical activity without purification.
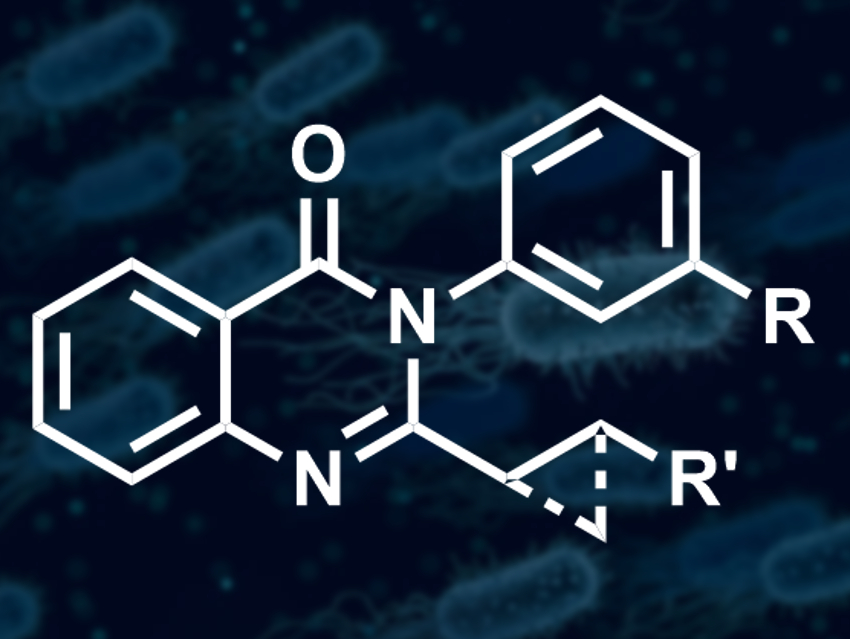
Alex J. O’Neill, Adam Nelson, and colleagues, University of Leeds, UK, have used ADS to expand a series of antibacterial quinazolinones (example pictured) that can kill multiple strains of Staphylococcus aureus. They set up an array of 220 reactions, using ten 2-iodophenylanilide or related compounds and 19 different anilines or related amines as substrates. Under Pd catalysis, these substrates can, in principle, form quinazolinones via carbonylation, N-substituted anilines, benzimidazoles, or quinolinones. The team carried out the array in well plates, using tri-tert-butylphosphine and tris(dibenzylideneacetone)dipalladium(0) as the catalyst system, molybdenum hexacarbonyl as a carbon monoxide source, and triethylamine as a base.
The activity of the unpurified products was tested using a standard method to assess antibacterial activity against a specific strain of S. aureus. The team found that six of the 220 reactions had produced active antibacterial compounds. These reactions were scaled up and the products purified using high-performance liquid chromatography (HPLC). In all cases, the team obtained quinazolinones with a hydrophobic substituent in the 2-position and a meta-substituted phenyl group in the 3-position (pictured).
The results provided insight into the structure–activity relationship (SAR) of the quinazolinones. The researchers found, for example, that the two-atom long linker at the 2-position was essential and could either be a CH2CH2 linker or a cyclopropane. They think that ADS may be useful for the discovery of other new antimicrobials that could avoid existing mechanisms of antibiotic resistance.
- Activity-directed expansion of a series of antibacterial agents,
Abbie Leggott, Justin E. Clarke, Shiao Chow, Stuart L. Warriner, Alex J. O’Neill, Adam Nelson,
Chem. Commun. 2020.
https://doi.org/10.1039/d0cc02361b