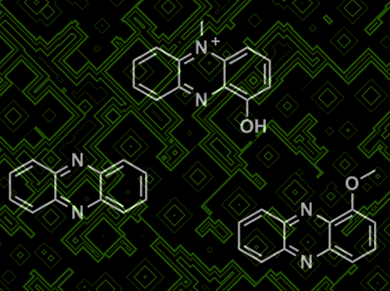
Screening mixtures of biologically active natural products (NP) for known compounds, and facilitating discovery of new compounds is very attractive. Screens, known as dereplication, used at an early stage help reduce the time, effort, and costs associated with identifying new compounds. Current methods analyze similarities in physical characteristics, but fail to provide exact compound structure information. However, by developing a robust database of MS/MS data from associated compounds, a data network can be formed, from cosine similarities, that allows known compounds and related analogues to be identified.
Pieter C. Dorrestein and colleagues from University of California, San Diego, USA, Chinese Academy of Sciences, China, and Universidade de Sao Paulo, Brazil, realized that MS/MS data gives a detailed fingerprint of a compound that allows this method to be precise. To test this technique, a number of marine and terrestrial microbial samples were tested and 58 molecules, including analogues, were correctly identified.
This method could be readily incorporated into NP discovery programs as well as databases compiled simply from MS/MS data of well-studied compounds and known standards.
- Molecular Networking as a Dereplication Strategy,
Jane Y. Yang, Laura M. Sanchez, Christopher M. Rath, Xueting Liu, Paul D. Boudreau, Nicole Bruns, Evgenia Glukhov, Anne Wodtke, Rafael de Felicio, Amanda Fenner, Weng Ruh Wong, Roger G. Linington, Lixin Zhang, Hosana M. Debonsi, William H. Gerwick, Pieter C. Dorrestein,
J. Nat. Prod. 2013.
DOI: 10.1021/np400413s