Bayram Terzi and Wiebke Teich, Ludwig‑Maximilians‑Universität München (LMU Munich), Germany, and their coauthors have presented the first photocaged or light-sensitive m⁷Gp₄G cap analogs (mRNA 5′ cap analogs). These compounds allow to control RNA translation and enzyme interactions by using light to remove the protective “cage,” thus activating the RNA molecule in a precise, time-dependent manner.
What did you do?
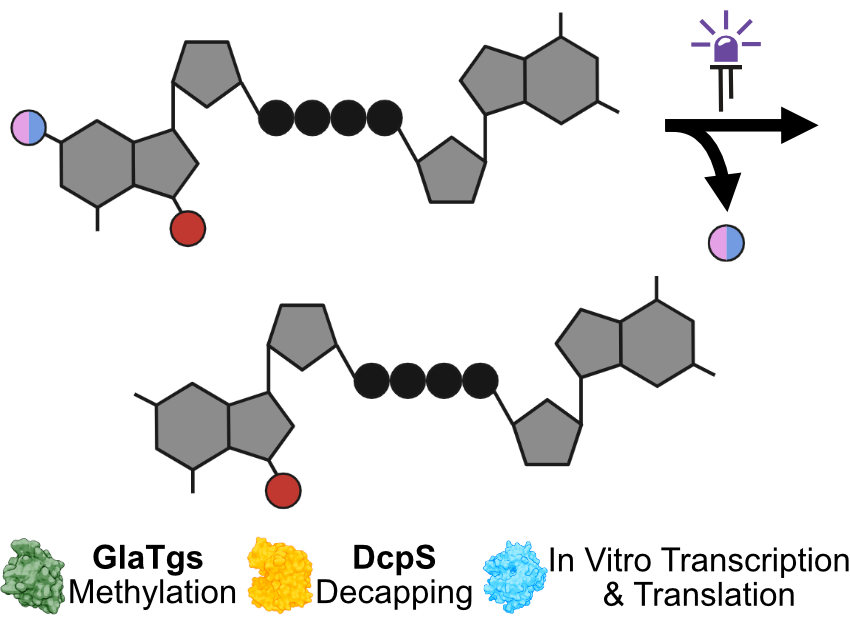
We chemically synthesized dinucleoside tetraphosphates (Np₄N) with two different nitrobenzyl-based photocleavable protecting groups.
While Np₄Ns are primarily found in bacteria, they are considered bacterial signaling molecules. However, adenosine tetraphosphate adenosine (Ap₄A)-capped mRNA was recently discovered in mammalian cells, although N7-methylated dinucleoside triphosphates (m⁷Gp₃N) remain the canonical 5′ cap structures of eukaryotic mRNA.
Using our photocaged tetraphosphate caps, we aimed to combine the m⁷Gp₄G structure with optochemical properties to enable temporal control over 5′ cap-specific interactions—namely, with RNA polymerase, methyltransferase, and decapping enzymes.
Why are you doing this?
The recent discovery of Ap₄A-capped eukaryotic mRNA aroused our interest in why Np₄N are incorporated into eukaryotic mRNA and how these mRNAs differ from canonically capped-mRNAs. Are Np₄N-capped mRNAs a side product of capping, or do they show yet unknown functions?
By studying their interactions with cap-specific enzymes in a light-controlled manner, we aim to advance the understanding of the differences and similarities between Np₄Ns and canonical 5′ cap structures.
What is new and cool about your study?
The photocaged tetraphosphate 5′ caps enable precise activation of 5′ cap interactions and translational processes by irradiation with light. Therefore, they enlarge our toolbox to not only study dinucleoside triphosphates, as we reported previously, but now investigate dinucleoside tetraphosphates.
What are your key findings?
We showed that our photocaged dinucleoside tetraphosphates are incorporated into mRNA and the additional phosphate within the phosphate bridge does not influence photolysis properties as well as recognition by 5′ cap-specific enzymes.
In contrast, photocleavable protecting groups at the N² position of m⁷G block all tested interactions, thereby enabling temporal control of 5′ cap interactions and translation.
What is the longer-term vision for your research?
We want to understand the role of Np₄Ns in bacterial and mammalian cells. Are they solely a side product, which is generated under cell stress or do they have a specific function? Why are they incorporated into mRNA?
We believe that our photocaged cap analogs are a helpful tool in analyzing the underlying mechanisms.
What part of your work was the most challenging?
The chemical synthesis of the dinucleoside tetraphosphates was challenging because the final coupling step is inefficient, and small impurities of GDP (arising from hydrolysis of GTP) favor the synthesis of dinucleoside triphosphates. Moreover, multiple purification steps are needed to ensure high purity of the 5′ cap analog.
Guanosine diphosphate (GDP) and guanosine triphosphate (GTP) are guanine-containing nucleotides involved in cellular energy transfer and signaling.
Thank you very much for sharing these insights.
The paper they talked about:
- Photocaged Dinucleoside Tetraphosphates for Light-Mediated Activation of 5′ Cap Interactions and Translation In Vitro,
Cedrik Kühling, Wiebke Teich, Bayram Terzi, Helena Schepers, Sabine Hüwel, Andrea Rentmeister,
ChemistryEurope 2025.
https://doi.org/10.1002/ceur.202500178

Bayram Terzi (left) and Wiebke Teich (right) are Ph.D. students in the laboratory of Professor Andrea Rentmeister at Ludwig‑Maximilians‑Universität München (LMU Munich), Germany.
Also of Interest

Just published articles from ChemistryEurope, the flagship journal of Chemistry Europe