Bacteria can rapidly develop a resistance to antibiotics, which is an important problem in healthcare. New antibiotics are urgently needed. RNA polymerase (RNAP) catalyzes the transcription of DNA into RNA, an essential process. Bacterial RNAP differs from that in eukaryotes, making it a useful target for antimicrobial drugs.
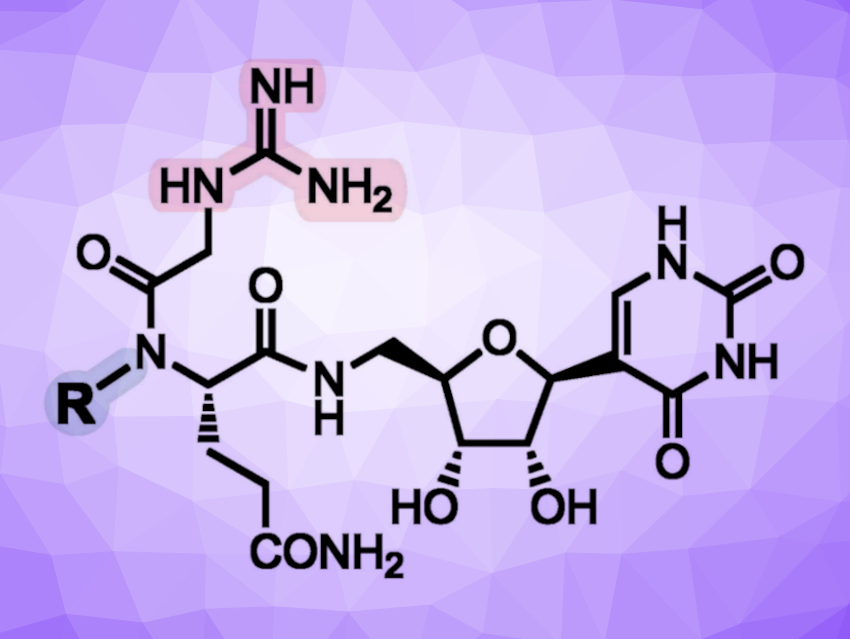
Pseudouridimycin (PUM) is a microbially produced C-nucleoside dipeptide antibiotic and a potent inhibitor of RNAP. Its structure contains guanidinylated glycine and hydroxyglutamine units, linked to a C-terminal β-pseudouridinyl amine. Despite the promising activity of PUM, its chemical instability limits the compound’s therapeutic potential—it undergoes relatively rapid decomposition in buffered aqueous solutions.
Juan R. Del Valle, University of Notre Dame, IN, USA, and colleagues have modified the dipeptide part of PUM to improve its stability. The primary decomposition pathway of PUM at physiological pH is a breaking of the hydroxamate bond mediated by the guanidine unit. Thus, the team’s strategies included modifying the hydroxamate unit (highlighted in blue above), replacing the guanidine unit with a less nucleophilic moiety (highlighted in red above), and introducing a conformational constraint into the guanidinylated glycine moiety by cyclization.
The stabilized analogues were evaluated for their RNAP inhibitory activity and antibacterial effects. A subset of analogues in which the labile hydroxamate was changed into a secondary or tertiary amide remained stable at pH 7.4 while maintaining antibacterial activity. These analogues are also more synthetically accessible than PUM, allowing for further optimization in the search for more potent drug candidates.
- Stabilizing Pseudouridimycin: Synthesis, RNA Polymerase Inhibitory Activity, and Antibacterial Activity of Dipeptide‐Modified Analogues,
Avraz F. Anwar, Christopher F. Cain, Michael J. Garza, David Degen, Richard H. Ebright, Juan R. Del Valle,
ChemMedChem 2023.
https://doi.org/10.1002/cmdc.202300474