Arnaud Gautier, CNRS, Université PSL, Paris, France, and colleague aimed to address the challenge of visualizing protein–protein interactions (PPIs) inside living cells at a resolution beyond the diffraction limit of light. Conventional fluorescence microscopy blurs fine details, making it difficult to observe how proteins assemble and interact at the nanoscale.
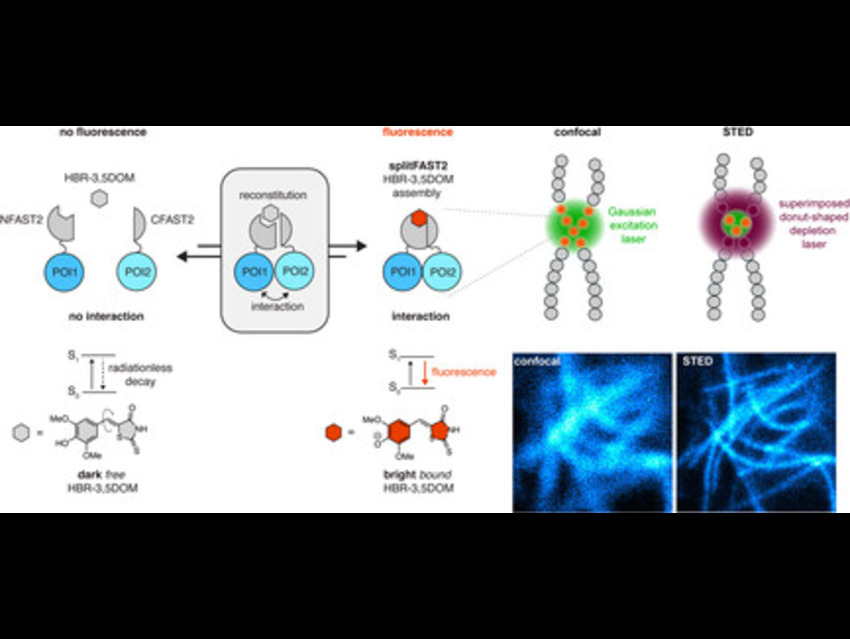
The team developed a chemogenetic split fluorescent reporter called splitFAST2, derived from FAST (Fluorescence-Activating and absorption-Shifting Tag), a small protein engineered into two fragments (NFAST2 and CFAST2). In simple terms, FAST is a protein that becomes fluorescent only when it binds a specific small-molecule dye (fluorogen). By splitting FAST into two pieces, the fragments alone remain non-fluorescent, but when they are brought together—such as by two proteins interacting—they reassemble into a functional fluorescent complex. This allows researchers to visualize protein–protein interactions in real time.
In their experiments, HeLa cells were transfected with DNA constructs encoding these fragments fused to proteins of interest: LifeAct, which binds actin filaments; Lamin, which forms the nuclear lamina; and Bcl-xL (B-cell lymphoma-extra large) fused to the BH3 domain, a short amino acid sequence that promotes programmed cell death (apoptosis). This strategy ensures that fluorescence appears only at sites where the proteins of interest interact, providing a highly specific readout.
Upon treatment with 10 µM of the synthetic fluorogen HBR-3,5DOM (4-hydroxybenzylidene rhodamine derivative), which binds the reconstituted FAST complex, bright fluorescence appeared specifically at interaction sites, minimizing background signals. Imaging with confocal and STED (stimulated emission depletion) microscopy, followed by image decorrelation analysis, showed that STED consistently enhanced resolution, reducing filament widths from approximately 300 nm to 90–170 nm.
This approach enabled precise visualization of dynamic protein–protein interactions and structural assemblies, including actin filaments, lamin oligomers, and Bcl-xL–BH3 complexes at mitochondria, demonstrating significant resolution gains and accurate localization in live cells. It opens the door to mapping PPIs and structural protein assemblies in livin gcells with unprecedented clarity and can be adapted to study signaling pathways, cytoskeletal remodeling, and apoptosis regulation.
Future directions include designing more splitFAST2-based probes for diverse proteins, improving fluorogen chemistry for brighter signals, and applying this method to disease models to uncover molecular mechanisms at the nanoscale.
- Super-Resolution Live-Cell Mapping of Protein–Protein Interactions Using Chemogenetic Split Reporters and Stimulated Emission Depletion Microscopy
Stephanie Board, Arnaud Gautier
ChemBioChem 2025
https://doi.org/10.1002/cbic.202500754