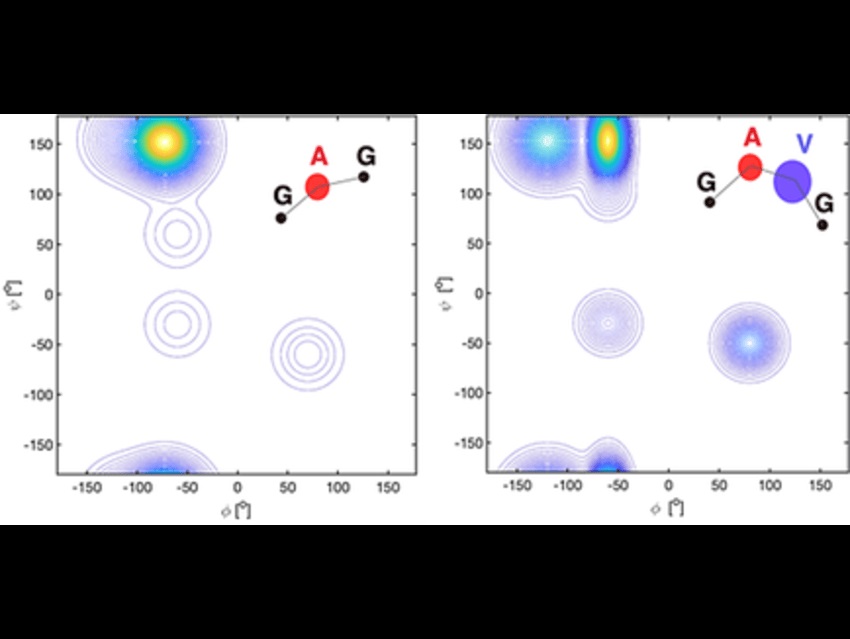
The study tackles a long-standing assumption in protein science: that unfolded and intrinsically disordered proteins (IDPs) behave like random coils. This view oversimplifies their actual behavior and ignores subtle structural preferences that influence biological function.
The research was led by Reinhard Schweitzer-Stenner from the Department of Chemistry, Drexel University, USA. The study highlights that short peptides, especially those containing alanine, can reveal local structural preferences in unfolded and disordered proteins. These preferences challenge the traditional random coil model and offer new benchmarks for molecular simulations, potentially improving force fields used in protein modeling. The study investigated the behavior of short peptides, using alanine dipeptides as simplified models to represent segments of proteins. Researchers employed a mix of experimental techniques, such as spectroscopy, along with computational simulations, specifically molecular dynamics using various force fields. A key analytical tool was the Ramachandran plot, which helped explore backbone conformations in detail. Simulations examined how solvent interactions and neighboring amino acids influence structural preferences, while comparisons across different force fields and water models evaluated how accurately these approaches mirrored experimental data.
The main finding was that alanine-rich peptides often adopt polyproline II-like conformations and do not behave randomly. This means that local structure matters, even in proteins that appear disordered overall. Thus, the current research shows that short peptides can act as valuable tools for improving molecular simulations and force fields, leading to better models of protein behavior. It deepens our understanding of intrinsically disordered proteins (IDPs), which, even without a stable structure, can still carry out important biological functions in Alzheimer’s; these insights may help guide future drug development. Looking ahead, further exploration of different amino acids and longer peptide chains could reveal more about protein folding and disorder.
- Local and Global Behavior of Unfolded and Intrinsically Disordered Peptides and Proteins
Reinhard Schweitzer-Stenner
ChemBioChem 2025
https://doi.org/10.1002/cbic.202500172