On the Precipice of a Signaling Renaissance
Optogenetics combines genetic and optical methods to cause or inhibit well-defined events in specific cells of living tissue. Opsin (a group of proteins made light-sensitive)-based optogenetics has revolutionized the field of neuroscience.
Investigators routinely express excitatory or inhibitory light-activated ion pumps or channels in a group of neurons and use light to induce or suppress action potentials. Optogenetic tools have been invaluable in allowing neuroscientists to pick apart neural circuits and correlate the activity of a collection of cells with animal behavior. The beauty and elegance of these studies are that light provides relatively non-invasive (compared to lesion studies), bio-orthogonal control with extreme spatial and temporal resolution relative to pharmacological and genetic studies.
However, cell biology, biochemistry, and signal transduction are still primarily studied using traditional methods. A variety of other photoreceptors are now being used to develop tools for controlling intracellular biochemistry and opening the door for biochemists to start to unravel the multidimensional processes of intracellular signaling.
Studying Subcellular Roles of Second Messenger Signaling Pathways
To give an example: Such tools are now available to study the subcellular roles of second messenger signaling pathways such as the cyclic adenosine monophosphate pathway (cAMP). cAMP is a derivative of adenosine triphosphate (ATP) and an important second messenger in many biological processes.
Traditional experiments treat cells with a pharmacological agonist of cAMP such as forskolin. Then, downstream activity of cAMP can be read via activity of a cAMP effector such as the cAMP-dependent kinase, PKA. However, cAMP has at least four different effectors in cells (PKA, exchange proteins activated by cAMP, cyclic nucleotide-gated ion channels, and popeye domain containing proteins). Each of these effectors is organized into discreet signaling nano- and microdomains.
Each branch of the cAMP-triggered pathway is extraordinarily complex. For example, PKA is sequestered to subcellular compartments via scaffolding proteins called A-Kinase Anchoring Proteins (AKAPs). cAMP can be generated either at the plasma membrane by GPCR coupled adenylate cyclases or in subcellular compartments by the pH- and Ca2+-dependent soluble adenylate cyclases. An additional complexity is caused by the phosphodiesterases present that hydrolyze cAMP, inactivating it, and generating four-dimensional gradients (see Fig. 1).
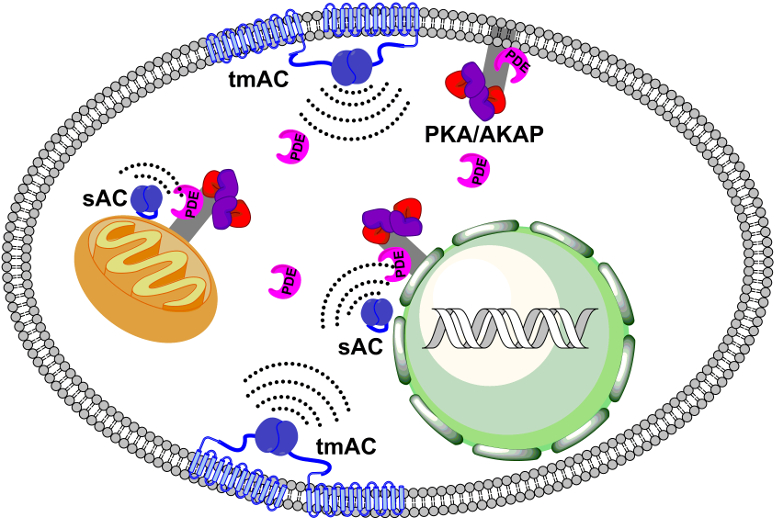
Figure 1. A schematic representation of cellular cAMP/PKA signaling organization (tmAC: Transmembrane adenylate cyclases, sAC: soluble adenylate cyclases, PDE: phosphodiesterases, PKA/AKAP: protein kinase A / A-kinase anchoring protein complexes).
It should be quite clear that the interpretation of the results from one experiment is complex given the variety of pathways that a single molecule can induce. Traditional methods for investigating intracellular biochemistry, such as genetic and pharmacological studies, have varying degrees of spatial or temporal resolution, respectively.
Recently, some breakthroughs have occurred that are just beginning to allow us to study individual cAMP/PKA signaling domains and even decouple cAMP from its downstream effector PKA, and thus, isolating these biochemical pathways. Work pioneered by Peter Hegemann and Georg Nagel [1] identified and characterized a small, blue light-activated adenylate cyclase. Since their initial work was published, many other photoactivated cyclases have been identified and characterized. These enzymes can potentially be directed to subcellular locations to control compartmentalized cAMP generation.
Our group recently published an optogenetic protein kinase A catalytic subunit, optoPKA [2]. It provides spatial and temporal control of PKA signaling at specific subcellular compartments. Taken together, these two tools offer a means to 1) decouple upstream cAMP generation from downstream PKA activity and 2) investigate the roles of ubiquitous signaling pathways at discrete cellular microdomains.
Outlook
The example of cAMP and PKA signaling is but one example of a wide variety of tools that are being developed and used by the biochemical community. From light-activated inhibitors of synaptic plasticity [3] to directional control of cell migration [4], these tools will surely revolutionize our understanding of the intricacies of cellular signaling and bring on a new age of in cellulo and in vivo biochemistry.
Just as opsin-based optogenetics are enabling our understanding of the complex web of neuronal circuitry, optogenetic tools for intracellular signaling will undoubtedly be a route to building a model of the inner workings of a cell.
Acknowledgments
1. Eshelman Institute for Innovation
2. The National Institutes of Health (1R01 NS103486-01A1)
References
[1] M. Stierl, P. Stumpf, D. Udwari, et al., Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa, J. Biol. Chem. 2011, 286, 1181–1188. https:doi.org/10.1074/jbc.M110.185496
[2] C. P. O’Banion, M. A. Priestman, R. M. Hughes, L. E. Herring, S. J. Capuzzi, D. S. Lawrence, Design and Profiling of a Subcellular Targeted Optogenetic cAMP-Dependent Protein Kinase, Cell Chem. Biol. 2018, 25, 100–109. https://doi.org/10.1016/j.chembiol.2017.09.011
[3] H. Murakoshi, M. E. Shin, P. Parra-Bueno, E. M. Szatmari, A. C. E. Shibata, R. Yasuda, Kinetics of Endogenous CaMKII Required for Synaptic Plasticity Revealed by Optogenetic Kinase Inhibitor, Neuron 2017, 94, 37–47. https://doi.org/10.1016/j.neuron.2017.02.036
[4] R. M. Hughes, D. S. Lawrence, Optogenetic engineering: light-directed cell motility, Angew Chem. Int. Ed. 2014, 53, 10904–10907. https://doi.org/10.1002/anie.201404198
For More Information
- Special Issue: Optogenetics,
ChemBioChem 2018, 19 (12). - Optogenetics: A Primer for Chemists,
Colin P. O’Banion, David S. Lawrence,
ChemBioChem 2018, 19, 1201–1216.
https://doi.org/10.1002/cbic.201800013