Lighting Up Your Proteins
There are about 500 naturally occurring amino acids, although a mere 20 of those are coded in our DNA as the building blocks of the proteins. Fundamentally though, there is an almost limitless number of hypothetical amino acids. Indeed, many synthetic amino acids have been synthesized for a wide range of applications.
It would be useful to find synthetic amino acids that could be substituted for their natural counterparts in a functioning protein to give said protein a new and useful extra property without disturbing its usual form and function. Unfortunately, finding and synthesizing such non-canonical amino acids is costly, and even when they can be produced, they often disrupt the target protein’s conformation and, thus, its functionality.
A New Canon for Amino Acids
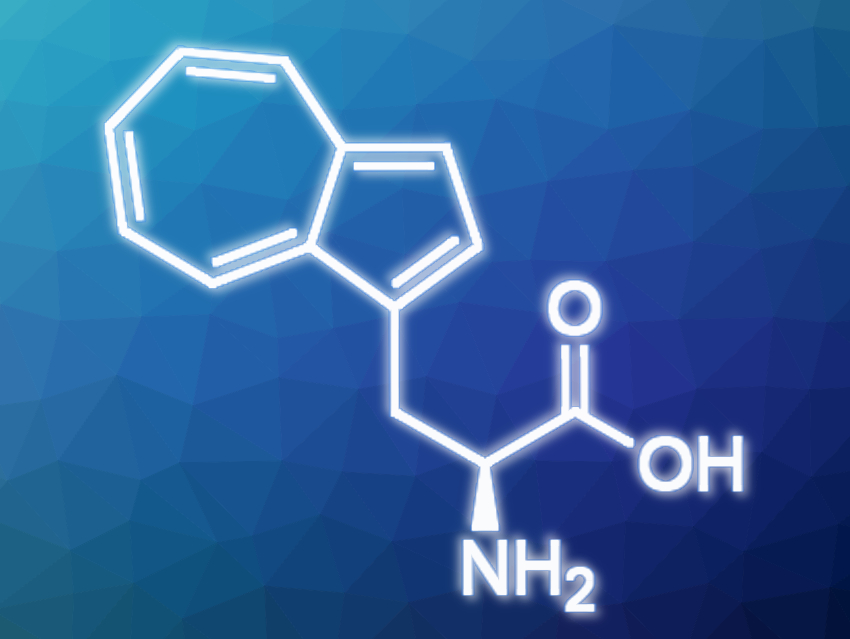
Frances H. Arnold, California Institute of Technology (CalTech), Pasadena, USA, and colleagues have found a new way to make the blue fluorescent non-canonical amino acid β-(1-azulenyl)-L-alanine (AzAla). The process involves a single enzymatic reaction step to give AzAla from readily available azulene and L-serine. The enzyme used for the production was an engineered tryptophan synthase that makes the target amino acid from the starting materials in a scalable manner. The product can be swapped into a protein to replace tryptophan without disrupting the conformation or the function of that protein. However, it still provides the benefit of adding a fluorescent marker to the protein that can be used as a tag for spectroscopic studies.
Such a tag might enable behavior and function to be investigated more closely, as well as facilitating studies of the interactions between proteins. The team explains that unlike the native tryptophan, their synthetic amino acid is insensitive to the environment in which it finds itself, spectroscopically speaking. It could be particularly suited to Förster resonance energy transfer (FRET) experiments for protein–protein interaction studies.
Fluorescence Excellence
Team member Patrick Almhjell explains: “AzAla is an excellent fluorescent probe that can very often be used to replace native tryptophan amino acids in proteins with minimal effect,” he told ChemViews Magazine. Because AzAla’s fluorescent properties are robust to environmental changes and relatively simple compared to those of tryptophan, it can be beneficial for experiments that require quantification based on fluorescent readout.
Similarly, AzAla can be used as an ultrafast heater, meaning that you can irradiate it with a certain wavelength of light and that energy will be efficiently converted to vibrational energy that can then be transferred to another nearby reporter and analyzed by researchers [1].
Adapting Amino Acids
The question is perhaps then whether or not the same enzymatic approach might work for other non-canonical amino acids. “We really pride ourselves in making enzymes that anybody can use,” Almhjell told us. He adds that “Creating the biocatalyst is very simple—because our TrpB variants are thermostable to around 90 °C or higher, we can express it in bacteria, then heat-treat the bacteria at 75 °C to denature the native bacteria proteins, pellet the denatured proteins and cell membrane via centrifugation, and you’re left with highly pure TrpB enzyme in the supernatant.”
For some of their reactions, the conditions have been optimized so that the non-canonical amino acid will precipitate directly out of solution at the end of the reaction. “All you have to do is collect it on filter paper and wash it,” adds Almhjell.
- Direct enzymatic synthesis of a deep‐blue fluorescent noncanonical amino acid from azulene and serine,
Ella J. Watkins, Patrick J. Almhjell, Frances H. Arnold,
ChemBioChem 2019.
https://doi.org/10.1002/cbic.201900497
Reference
- [1] Site-Resolved Observation of Vibrational Energy Transfer Using a Genetically Encoded Ultrafast Heater,
Tobias Baumann, Matthias Hauf, Fabian Schildhauer, Katharina B. Eberl, Patrick M. Durkin, Erhan Deniz, Jan G. Löffler, Carlos G. Acevedo-Rocha, Jelena Jaric, Berta M. Martins, Holger Dobbek, Jens Bredenbeck, Nediljko Budisa,
Angew. Chem. Int. Ed. 2019, 58, 2899–2903.
https://doi.org/10.1002/anie.201812995