There is not longer just one type of SARS-CoV-2 virus. New variants continue to emerge as a result of mutations. This raises the question of whether recovered individuals are still protected from reinfection or whether current vaccines are losing effectiveness. All currently used vaccines prepare our immune system for viral infection by creating antibodies against the spike protein of the original first-wave virus. Most mutations of SARS-CoV-2 do not have a noticeable impact on us, but some do.
To reduce confusion and avoid geographic stigmas, the World Health Organization (WHO) unveiled a new nomenclature for SARS-CoV-2 viral variants in May 2021. Variants that the WHO classifies as “variant of concern” (VOC) or “variant of interest – variant under observation” (VOI) are named after letters of the Greek alphabet. These abbreviations do not replace the scientific names but serve the layperson.
Below, we have a closer look at the variants of concern (VOC) that have been designated by the WHO (as of July 2, 2021). This means variants of the SARS-CoV-2 virus that show (i) an increase in transmissibility or detrimental change in COVID-19 epidemiology, or (ii) an increase in virulence or a change in clinical disease presentation, or (iii) a decrease in effectiveness of public health and social measures or available diagnostics, vaccines, therapeutics.
1 The SARS-CoV-2 Coronavirus
.png)
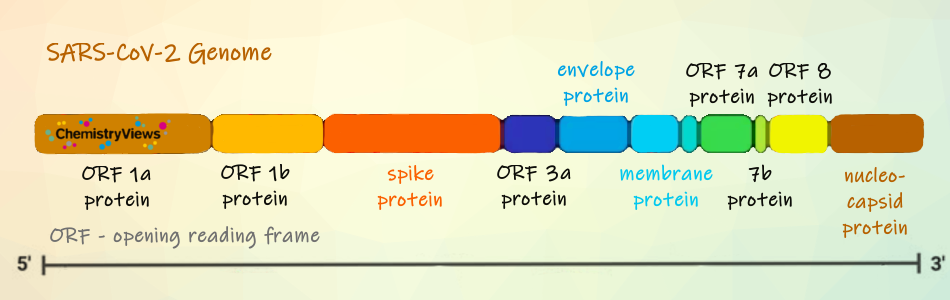
Coronaviruses (CoVs) are a group of enveloped viruses with a positive single-stranded RNA genome. They have the largest RNA viral genome. SARS-CoV-2 contains four structural proteins: the spike, envelope, membrane, and nucleocapsid proteins. CoVs rely on their spike proteins for entering the host cells (see Coronavirus Entering and Replicating in a Host Cell).
In the following we will look at the spike protein. It binds to the host receptor through the receptor-binding domain (RBD) in the S1 subunit, followed by the fusion of the S2 subunit to the host cell membrane (see below).
.png)
2 Coronavirus Variants of Concern (VOC)
|
WHO Label |
Pango Lineage |
Spotted in |
Number of Muations in Spike |
Other Names |
||
|
alpha |
B.1.1.7. |
September 2020 in UK |
8 |
GR/501Y.V1 GRY (formerly) |
||
|
beta |
B.1.351 B.1.351.2 |
May 2020 in South Africa |
9 |
GH/501Y.V2 |
||
|
gamma |
P.1 P.1.1 P.1.2 |
November 2020 in Brazil |
12 |
GR/501Y.V3 |
||
|
delta |
B.1.617.2 AY.1 AY.2 |
October 2020 in India |
7 |
G/4478K.V1 |
||
|
omicron (includes BA.1, BA.2, BA.3, BA.4, BA.5) |
B.1.1.529
|
November 2021 in Botswana and other countries |
>30 |
GR/484A |
.png)
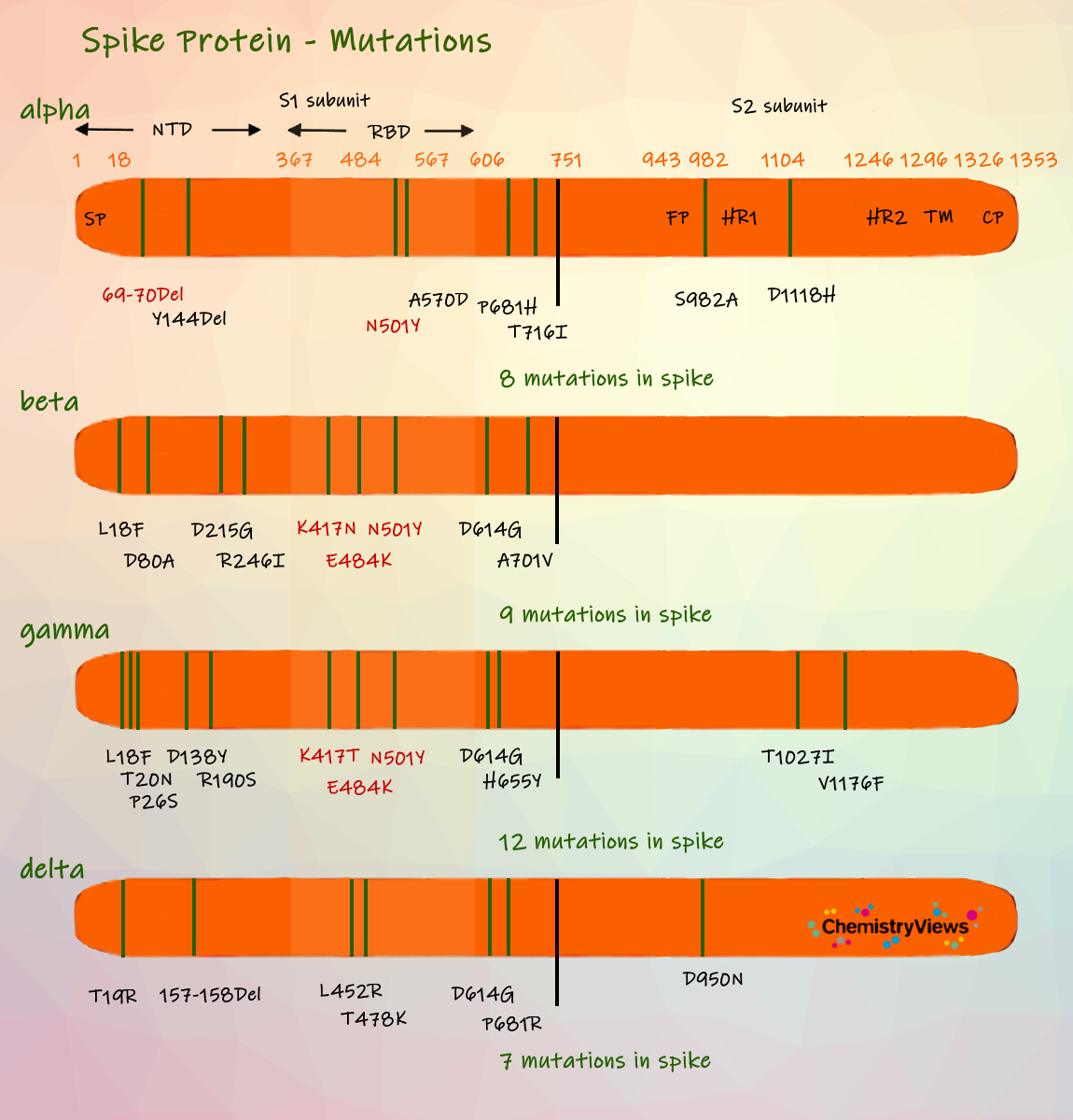
Swaps or deletions of single amino acids can change the shapes of proteins, and thus, the properties of the virus. Researchers are particularly concerned about mutations in the N-terminal domain of the spike protein (some antibodies target it) and the receptor-binding domain (RBD) of the spike protein (as mentioned earlier, it docks to the ACE2 receptors on human cells and initiates the infection process). Mutations that are particularly worrisome are shown in red in the picture above.
The table below lists which effect selected mutations have for the protein and the virus and in which variants they occure.
|
Spike Mutation |
Virus Variant |
Effect |
|
Δ69-70 (69-70del) |
alpha |
may partially interfere with RT-PCR testing, |
|
Y144Del |
alpha |
apparently alters the shape of the spike protein so that it can escape some antibodies |
|
K417N |
beta, gamma |
key epitope for binding neutralizing antibodies => humoral response can be bypassed |
|
L452R |
delta |
in vitro higher ACE2 receptor affinity, enhanced infectivity, alteration of antigenic properties (affects the binding of certain monoclonal antibodies and could, thus, lead to immune escape) |
|
E484K |
alpha (partly), beta, gamma |
increased binding to human ACE2 receptor => could be responsible for the virus no longer being recognized by antibodies |
|
N501Y |
alpha, beta, gamma |
stronger binding affinity to human ACE2 receptor => increased transmissibility |
|
D614G |
beta, gamma, delta |
more open conformation of the spike protein => better binding to ACE2 receptor protein => higher in vitro infectivity |
|
P681H |
alpha |
may play a role in spike protein processing |
|
Virus Variant |
Effect Compared to Original Virus/Wild-Type |
|
alpha |
relative transmissibility about 50 % higher, effectiveness of vaccines only slightly weaker |
|
beta |
up to 50 % more infectious, possible reduction of vaccine effectiveness |
|
gamma |
increased transmissibility, possible reduction of vaccine effectiveness |
|
delta |
most transmissible form, becoming the world’s dominant strain reduction of vaccine effectiveness |
|
omicron |
Preliminary evidence suggests: |
References
- Tracking SARS-CoV-2 Variants, World Health Organization (WHO) (accessed July 1, 2021)
- Isabelle Viktoria Maucher, Übersicht der Corona-Mutanten, Gelbe Liste Online 2021. (accessed June 17, 2021; in German)
- Effectiveness of COVID-19 vaccines against the B.1.617.2 variant,
Jamie Lopez Bernal, Nick Andrews, Charlotte Gower, Eileen Gallagher, Ruth Simmons, Simon Thelwall, Julia Stowe, Elise Tessier, Natalie Groves, Gavin Dabrera, Richard Myers, Colin Campbell, Gayatri Amirthalingam, Matt Edmunds, Maria Zambon, Kevin Brown, Susan Hopkins, Meera Chand, Mary Ramsay,
medRxiv 2021.
https://doi.org/10.1101/2021.05.22.21257658 - Safety and Immunogenicity Report from the Com-COV Study – a Single-Blind Randomised Non-Inferiority Trial Comparing Heterologous And Homologous Prime-Boost Schedules with An Adenoviral Vectored and mRNA COVID-19 Vaccine,
Xinxue Liu, Robert H. Shaw, Arabella SV Stuart, Melanie Greenland, Tanya Dinesh, Samuel Provstgaard-Morys, Elizabeth Clutterbuck, Maheshi N. Ramasamy, Parvinder K. Aley, Yama Farooq Mujadidi, Fei Long, Emma Plested, Hannah Robinson, Nisha Singh, Laura L. Walker, Rachel White, Nick Andrews, J. Claire Cameron, Andrea M. Collins, Daniela M. Ferreira, Helen C. Hill, Christopher A. Green, Bassam Hallis, Paul T. Heath, Saul N. Faust, Adam Finn, Teresa Lambe, Rajeka Lazarus, Vincenzo Libri, Mary E. Ramsay, Robert C. Read, David P. J. Turner, Paul J. Turner, Jonathan S. Nguyen-Van-Tam, Matthew D. Snape, Com-COV Study Group,
SSRN Electronic Journal 2021.
https://doi.org/10.2139/ssrn.3874014 - Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1-nCoV19 and BNT162b2: a prospective cohort study,
David Hillus, Tatjana Schwarz, Pinkus Tober-Lau, Hana Hastor, Charlotte Thibeault, Stefanie Kasper, Elisa T. Helbig, Lena J. Lippert, Patricia Tscheak, Marie Luisa Schmidt, Johanna Riege, André Solarek, Christof von Kalle, Chantip Dang-Heine, Piotr Kopankiewicz, Norbert Suttorp, Christian Drosten, Harald Bias, Joachim Seybold, EICOV/COVIM Study Group, Claudia Conrad, Doris Steuer, Ute Gläser, Anne-Sophie Sinnigen, Carolin Rubisch, Nadine Olk, Lisbeth Hasler, Angela Sanchez-Rezza, Paolo Kronenberg, Alexandra Horn, Willi Koch, Paula Stubbemann, Julie-Anne Gabelich, Friederike Münn, Julia Tesch, Petra Mackeldanz, Leon Bergfeld, Tobias Bleicker, Jörn Ilmo Beheim-Schwarzbach, Anna Hiller, Sophia Brumhard, Lara Bardtke, Kai Pohl, Daniel Wendisch, Philipp Georg, Denise Treue, Dana Briesemeister, Jenny Schlesinger, Andreas Hetey, Luisa Kegel, Annelie Richter, Ben Al-Rim, Birgit Maeß, Kerstin Behn, Michelle Lysi, Saskia Zvorc, Maria Rönnefarth, Sein Schmidt, Alexander Krannich, Isabelle Schellenberger, Georg Schwanitz, Viktoria Schenkel, Norma Bethke, Claudia Hülso, Sebastian Dieckmann, Christian Peiser, Florian Kurth, Victor Max Corman, Leif Erik Sander,
medRxiv 2021.
https://doi.org/10.1101/2021.05.19.21257334
Also of Interest
- Clever Picture: Comparing COVID-19 Vaccines
Four Covid-19 vaccines have been approved in the EU to date
https://doi.org/10.1002/chemv.202100033 - Searching for a Vaccine
Picture describes what happens during vaccination and gives a snapshot of which Covid-19 vaccine candidates are in which clinical phases
https://doi.org/10.1002/chemv.202000075 - Interview: Interview with Ingmar Hoerr, Pioneer of mRNA Technology
As a Ph.D. student, Hoerr discovered that mRNA can be used as a therapeutic vaccine or agent when administered directly into tissues
https://doi.org/10.1002/chemv.202100030 - ChemistryViews Collection: SARS-CoV-2 Virus – What We Know About Vaccines and Immunity
Update 26.11.2021
B.1.1.529 as new variant of concern added