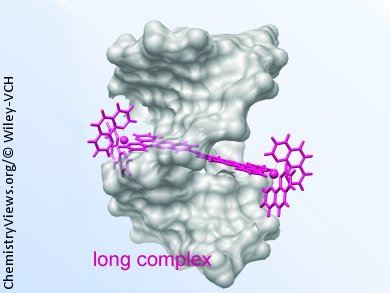
Since the discovery that DNA is the template for protein synthesis, efforts have been made to develop DNA binding drugs. One of the main challenges for their development is achieving specificity, while maintaining biological activity. Threading intercalation is an unusual DNA binding mode observed for some dumbbell-shaped molecules that consist of a flat heteroaromatic ring system with bulky and often charged substituents at both ends. Thus, one bulky substituent is threaded through the base-pair stack, intercalating the flat aromatic section between the bases with the bulky substituents each located in a groove in the bound state. Achieving this intercalation requires large distortions of the DNA structure, resulting in the threading and unthreading being extremely slow processes. Several reports suggest a correlation between slow dissociation kinetics and biological activity for DNA binding drugs, making intercalating compounds therapeutically interesting.
Per Lincoln and co-workers, Chalmers University of Technology, Gothenburg, Sweden, have investigated the effect of the bridging ligand length of binuclear ruthenium threading intercalators on their DNA binding properties. They have found that elongation of the bridging ligand in binuclear ruthenium threading intercalators increases the binding affinity by increasing the association rate and decreasing the dissociation rate, but decreases the selectivity for AT-rich DNA. The bridging ligand length also controls the enantioselectivity of the threading interaction, with an increasing preference for the ΔΔ enantiomer as the distance between the ruthenium centers increases.
These findings are important for the design of new DNA binding drugs with slow kinetics.
- Bridging Ligand Length Controls AT Selectivity and Enantioselectivity of Binuclear Ruthenium Threading Intercalators,
Johan R. Johansson, Yubo Wang, Mattias P. Eng, Nina Kann, Per Lincoln, Johanna Andersson,
Chem. Eur. J. 2013.
DOI: 10.1002/chem.201300483