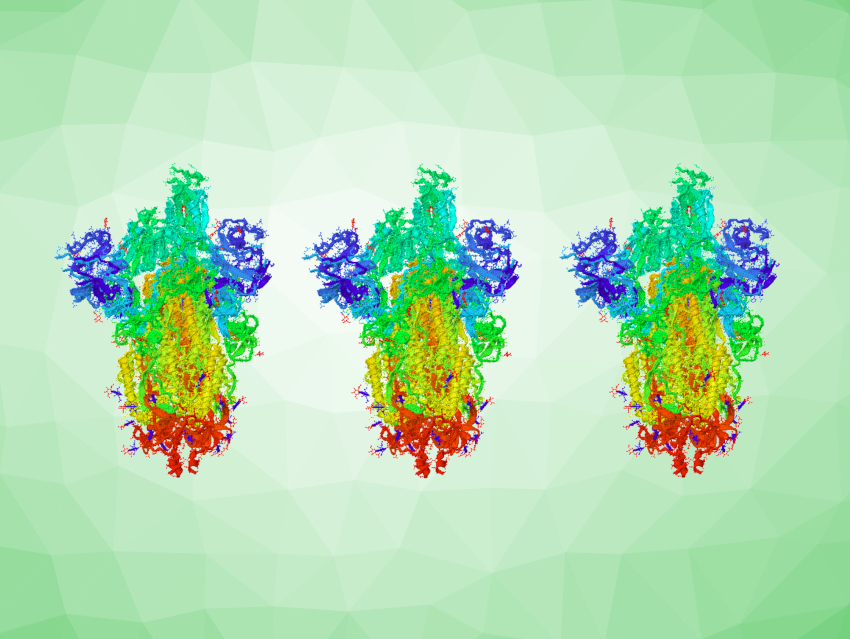
New variants of SARS-CoV-2 keep emerging during the pandemic. The omicron variant has been found concerning due to its high transmissibility and potential immune escape properties, and it has become dominant in many countries. Understanding how vaccinations and previous infections affect the protection against omicron and other variants of concern is important.
Oliver T. Keppler, Ludwig-Maximilians-Universität (LMU) München and German Centre for Infection Research (DZIF), both Munich, Germany, Ulrike Protzer, DZIF and Technical University of Munich (TUM), Germany, and colleagues have performed a longitudinal study to investigate neutralizing antibody dynamics in both vaccinated and recovered subjects. The researchers identified individuals who had contracted SARS-CoV-2 during the first wave of the pandemic in spring 2020 and added a matched second group of people who had not been infected. The study included 98 recovered persons and 73 persons without prior infection. Both groups were offered vaccination with the mRNA-based BioNTech/Pfizer COVID-19 vaccine and their antibody responses were measured. The participants were continuously followed from spring 2020 to the last quarter of 2021.
The team found that three exposures to the SARS-CoV-2 spike protein lead to the production of virus-neutralizing antibodies not only in high quantities, but also in high quality. These high-quality antibodies showed superior neutralizing activity against all variants of concern, including the omicron variant. This applies to various constellations of exposures: triple-vaccinated persons, persons who had recovered from COVID-19 and then had two vaccinations, and double-vaccinated persons who then had a breakthrough infection.
- Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern,
Paul R. Wratil, Marcel Stern, Alina Priller, Annika Willmann, Giovanni Almanzar, Emanuel Vogel, Martin Feuerherd, Cho-Chin Cheng, Sarah Yazici, Catharina Christa, Samuel Jeske, Gaia Lupoli, Tim Vogt, Manuel Albanese, Ernesto Mejías-Pérez, Stefan Bauernfried, Natalia Graf, Hrvoje Mijocevic, Martin Vu, Kathrin Tinnefeld, Jochen Wettengel, Dieter Hoffmann, Maximilian Muenchhoff, Christopher Daechert, Helga Mairhofer, Stefan Krebs, Volker Fingerle, Alexander Graf, Philipp Steininger, Helmut Blum, Veit Hornung, Bernhard Liebl, Klaus Überla, Martina Prelog, Percy Knolle, Oliver T. Keppler, Ulrike Protzer,
Nat. Med. 2022.
https://doi.org/10.1038/s41591-022-01715-4
Also of Interest
- Collection: SARS-CoV-2 Virus
What we know about the new coronavirus and COVID-19