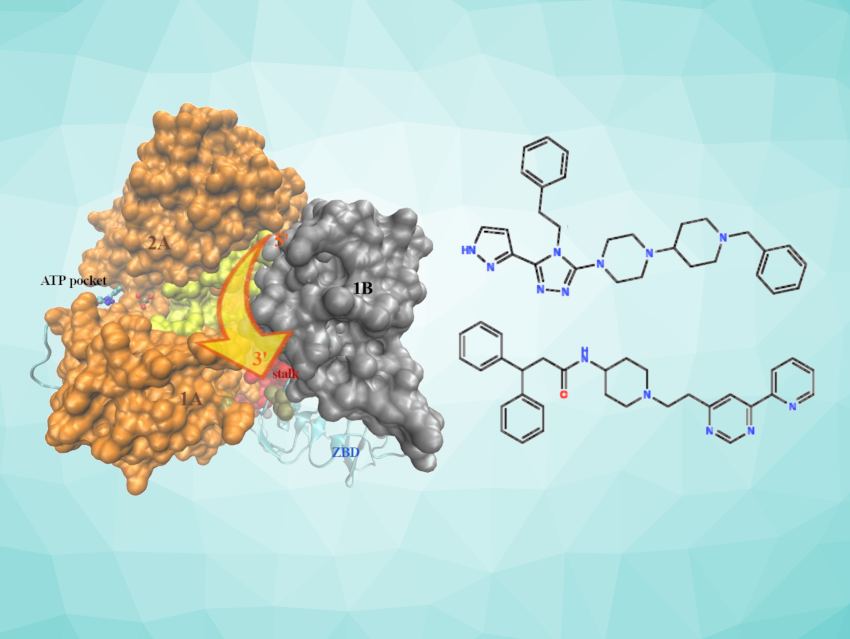
In the wake of the COVID-19 pandemic, new antiviral agents to cure coronavirus-related infections are interesting research targets. The NSP13 helicase, an enzyme that catalyzes the unwinding of viral RNA in virus replication, could be a promising target for designing broad-spectrum antiviral drugs. Inhibition of this protein could be achieved by designing compounds that block the entrance or exit of the central RNA channel, a structural motif that is highly conserved (i.e., very similar) across coronaviruses.
Piero Procacci, Università degli Studi di Firenze, Sesto Fiorentino, Italy, and colleagues have developed a “computational pipeline” that can be used to identify possible inhibitors of the SARS-CoV-2 NSP13 helicase (pictured). The team’s approach is based on the preliminary generation of potential NSP13 helicase binders using machine learning (ML) techniques based on generative neural networks, followed by an extensive docking campaign. Docking is a method in molecular modeling that can be used to predict the configuration in which a ligand and a target bind to each other to form a complex, as well as the resulting binding affinity. The potency of the candidate compounds was estimated using four popular docking scoring functions.
The best “hits” were further screened using accurate simulations on a high-performing parallel computing cluster. Using this approach, the researchers identified two compounds (pictured) with particularly strong predicted activities. These compounds could potentially be good candidates for the development of a broad-spectrum drug for coronavirus infections. According to the team, they are currently being synthesized and tested experimentally.
- Identification of Potential inhibitors of the SARS‐CoV‐2 NSP13 Helicase via Structure‐Based Ligand Design, Molecular Docking and Nonequilibrium Alchemical Simulations,
Giorgio Di Paco, Marina Macchiagodena, Piero Procacci,
ChemMedChem 2024.
https://doi.org/10.1002/cmdc.202400095