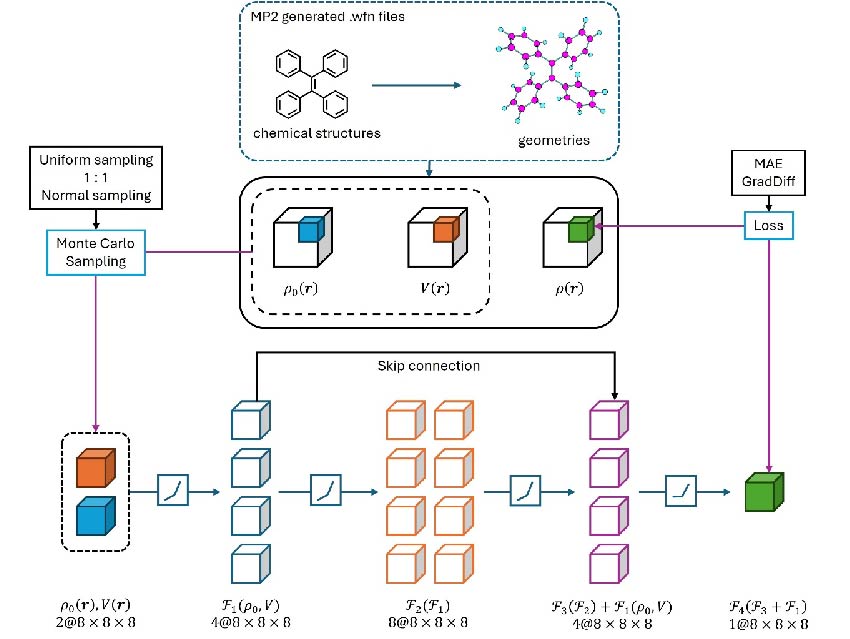
In quantum chemistry, accurately predicting electron density distributions for large systems has long faced dual challenges: traditional methods demand excessive computational resources, while machine learning typically requires tens of thousands of labeled data points. A breakthrough study by Ben Zhong Tang’s team at The Chinese University of Hong Kong, Shenzhen (CUHK-Shenzhen) and Junyi Gong’s team at Shenzhen MSU-BIT University introduces GED-CRN (Grid-sampled Electron Density Convolutional Residual Network), achieving near-second-order Møller–Plesset perturbation theory (MP2) accuracy (test set MAE <3.0×10⁻⁴ bohr⁻³) with only 19 training molecules—reducing computation from hours/days to seconds. MP2 is a widely used post-Hartree–Fock method for including electron correlation.
The researchers addressed the “data poverty in abundance” paradox in materials science by developing a physics-informed deep learning approach. Their cubic sampling strategy divides molecular space into 2×2×2 bohr³ units (containing 8×8×8 grid points), generating thousands of effective samples per molecule. The dual-channel 3D CNN (three-dimensional convolutional neural network) architecture incorporates both preliminary electron density and nuclear potential fields, while the novel GradDiff (Gradient Difference) loss function enables simultaneous prediction of density and spatial gradients.
Remarkably, GED-CRN shows enhanced performance for AIE (aggregation-induced emission) systems (50% lower error than conventional molecules) and achieves <0.5% error in key quantum chemical properties. The open-source model enables second-scale predictions, offering a 1000× speedup over MP2 for large-scale virtual screening of functional materials.
- GED-CRN Breaks the Data Barrier: High-Fidelity Electron Density Prediction Using Only 19 Training Molecules
Junyi Gong, Haoran Wang, Zijie Qiu, Zheng Zhao, Ben Zhong Tang
Aggregate 2025
https://doi.org/10.1002/agt2.70119