Protein misfolding and aggregation are characteristic of different neurodegenerative disorders. For example, amyloid-β peptide (Aβ) oligomers are considered the primary neurotoxic species relevant to Alzheimer’s disease, the most common neurodegenerative illness associated with dementia. However, the exact structures of Aβ oligomers are difficult to determine due to their high heterogeneity and metastability. The development of new methods that precisely capture single Aβ oligomers from mixtures could be useful for elucidating the structural reasons behind oligomer toxicity in Alzheimer’s disease.
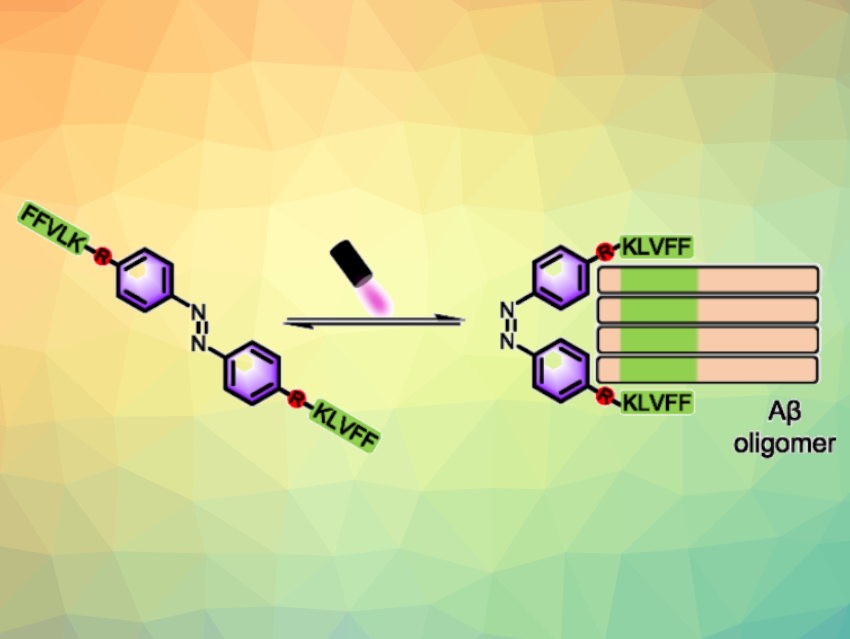
Xiaohui Wang, Nanjing Tech University, China, Xiaoyong Wang, Nanjing University, China, and colleagues have developed a general method for precisely capturing specific Aβ oligomers in vitro and in vivo using light-controlled molecular tweezers (LMTs). The LMTs are composed of two Aβ-targeting pentapeptides (with the sequence KLVFF, or Lys-Leu-Val-Phe-Phe) and a photoswitchable azobenzene core (pictured). The inactive form has a trans configuration. Light irradiation can activate LMTs to form a tweezer-like cis configuration that preferentially binds to specific oligomers via multivalent interactions of the LMTs’ KLVFF units and the Aβ oligomers.
As a proof of concept, cis-LMT1 can precisely capture and stabilize dimers of a synthetic Aβ from an Aβ mixture in vitro, as well as a natural Aβ in transgenic Caenorhabditis elegans in vivo. The LMTs could be promising starting scaffolds to achieve the extraction or elimination of specific natural Aβ oligomers via a reversible photoisomerization. This could be useful for structure and function studies and for the development of oligomer-targeted therapies. The strategy could potentially also be extended for the development of other molecular tweezers for capturing different protein amyloid oligomers of interest.
- Light‐controlled molecular tweezers capture specific amyloid oligomers,
Chengyuan Qian, Jiefang Chen, Cheng Wang, Qiang Wang, Xiaoyong Wang, Xiaohui Wang,
Aggregate 2023.
https://doi.org/10.1002/agt2.463