Coronaviruses have glycoprotein “spikes” on their surface, which help the virus to enter host cells by fusing the virus and the cell membrane. The structure of the spike protein of the coronavirus SARS-CoV-2 that causes the current outbreak of COVID-19 has recently been determined by cryogenic electron microscopy (cryo-EM). Cryo-EM uses very low temperatures to allow the determination of biomolecular structures at almost atomic resolution.
The coronavirus spike contains a receptor-binding domain (RBD) subunit, which binds to a receptor on the surface of the host cells’ membrane—the angiotensin-converting enzyme 2 (ACE2). However, the full-length structure of ACE2 has been difficult to determine so far.
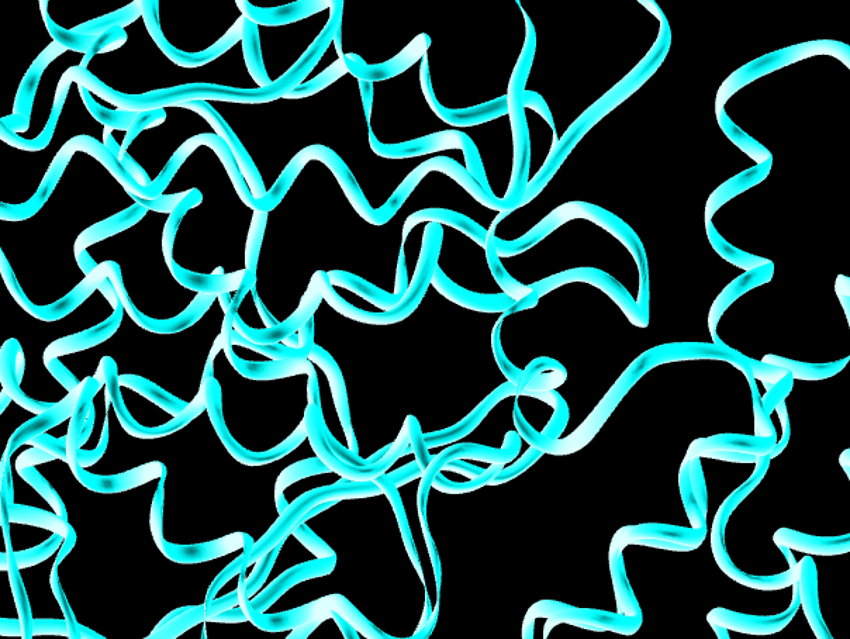
Qiang Zhou, Key Westlake Institute for Advanced Study and Westlake University, both, Hangzhou, China, have obtained cryo-EM structures of full-length human ACE2. The measurements were performed in the presence of the neutral amino acid transporter B0AT1, which provides stabilization. The structures were recorded both with and without the RBD of the spike protein of SARS-CoV-2. The team co-expressed the ACE2 and B0AT1 proteins in a human cell line, mixed a part of the samples with RBD, and studied the resulting complexes using cryo-EM. The structures have an overall resolution of 2.9 Å and a local resolution of 3.5 Å at the ACE2–RBD interface.
The researchers found that the ACE2–B0AT1 complex has a dimeric structure, with the ACE2 dimer sandwiched by B0AT1. Overall, ACE2 consists of a peptidase domain (PD) on the outer side of the cell membrane and a collectrin-like domain (CLD) that ends with a single transmembrane helix and an intracellular segment. The structure of the ternary RBD–ACE2–B0AT1 complex shows that each PD binds to one RBD, mainly via polar interactions. This is similar to the interaction between the SARS virus and ACE2. According to the team, these results could be helpful for the structure-based design of compounds that bind to either ACE2 or the spike protein of coronaviruses and, thus, suppress viral infection.
- Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2,
Renhong Yan, Yuanyuan Zhang, Yaning Li, Lu Xia, Yingying Guo, Qiang Zhou,
Science 2020.
https://doi.org/10.1126/science.abb2762
Also of Interest
- α-Ketoamides Keep Different Viruses from Multiplying,
ChemistryViews.org 2020.
New broad-spectrum antivirals against coronaviruses and enteroviruses - Clever Picture: Coronavirus Entering and Replicating in a Host Cell,
Vera Koester,
ChemViews Mag. 2020.
https://doi.org/10.1002/chemv.202000018
Where the coronavirus comes from and how it infects the human body - Structure of “Spike” Protein in New Coronavirus Determined,
ChemistryViews.org 2020.
Results could help to develop a vaccine and antibody treatments - Antiviral Drug Shows Promise Against MERS Coronavirus,
ChemistryViews.org 2020.
The broad-acting drug remdesivir could also be useful against the current coronavirus disease (COVID-19) - How Long Can Coronaviruses Live on Surfaces?,
ChemistryViews.org 2020.
Literature review finds coronaviruses could persist on hard surfaces for up to nine days, but can be killed with the right disinfectant - Suggesting Potential Drug Candidates for Coronavirus,
ChemViews Mag. 2020.
Summarizing the current research and suggesting potential drug candidates for treating patients suffering from the 2019-nCoV acute respiratory disease
- LitCovid
Curated literature hub for tracking up-to-date scientific information about COVID-19 - Many publishers and other entities have signed a joint statement to ensure that COVID-19 research findings and data are shared rapidly and openly