The coronavirus SARS-CoV-2 causes the current outbreak of the respiratory disease COVID-19. Antiviral medication to treat COVID-19 is urgently needed. A possible drug target in coronaviruses is the main protease (Mpro also called 3CLpro), an enzyme that is essential for replication of the virus.
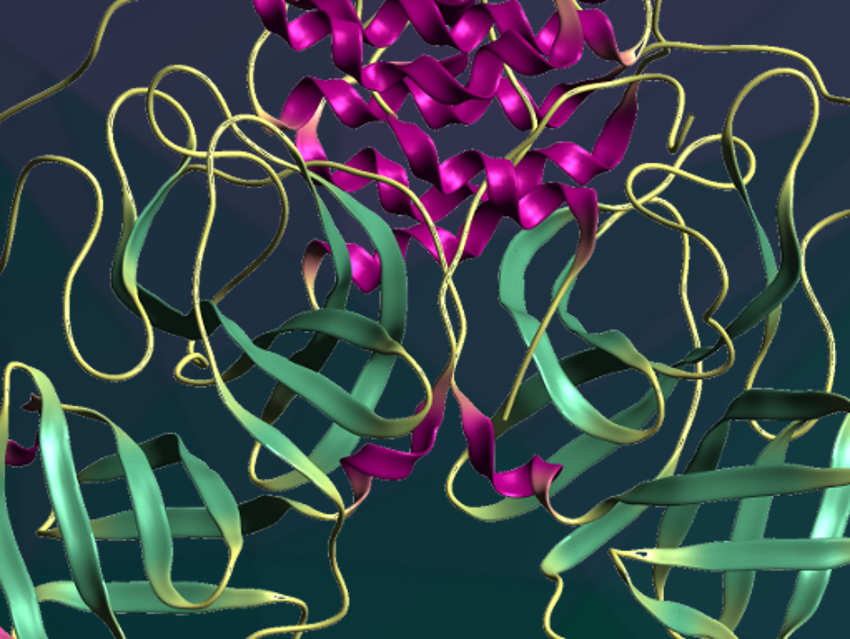
Rolf Hilgenfeld, University of Lübeck, Germany, and colleagues have recently designed and synthesized peptidomimetic α-ketoamides to be used as inhibitors of the main proteases of coronaviruses. To allow the design of improved α-ketoamide inhibitors, the researchers have determined the crystal structure of the Mpro of SARS-CoV-2 at a resolution of 1.75 Å using X-ray crystallography.
The structure is very similar to the Mpro of the related SARS virus. It is a dimer that features six-stranded antiparallel β-barrels, with the substrate-binding site between them, and a cluster of five helices involved in dimerization of the enzyme. The team used the crystal structure to investigate the interactions of Mpro with α-ketoamides and optimize the substituents of the drug candidates. They obtained a potent inhibitor of the SARS-CoV-2 Mpro. The optimized drug candidate showed promising properties in a mouse study: It remains in the lung tissue for some time and could potentially be administered directly by inhalation.
- Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors,
Linlin Zhang, Daizong Lin, Xinyuanyuan Sun, Ute Curth, Christian Drosten, Lucie Sauerhering, Stephan Becker, Katharina Rox, Rolf Hilgenfeld,
Science 2020.
https://doi.org/10.1126/science.abb3405
Also of Interest
- α-Ketoamides Keep Different Viruses from Multiplying,
New broad-spectrum antivirals against coronaviruses and enteroviruses - Clever Picture: Coronavirus Entering and Replicating in a Host Cell,
Vera Koester,
https://doi.org/10.1002/chemv.202000018
Where the coronavirus comes from and how it infects the human body - Collection: SARS-CoV-2 Virus
What we know about the coronavirus and COVID-19 – all articles related to the coronavirus published on ChemistryViews