SARS-CoV-2, the coronavirus that causes the COVID-19 pandemic, causes different symptoms to different individuals. Many reasons have been given. Shiyue Fang, Michigan Technological University, Houghton, USA, and colleagues propose that an individual’s epitranscriptomic system could be responsible as well.
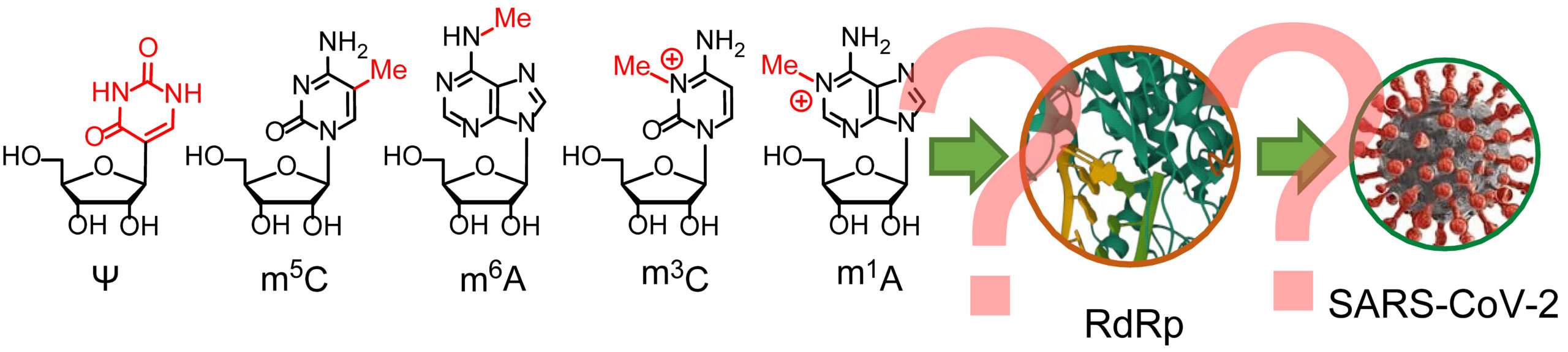
The epitranscriptomic system refers to the set of modifications that can be added to RNA molecules, including but not limited to modifications of nucleotides such as methylation, acetylation, and pseudouridylation. These modifications can alter RNA stability, structure, and function, and can have a significant impact on gene expression and various cellular processes. It has been found that the system can also work on viral RNAs, and thus may affect events in viral life cycles including viral RNA synthesis.
The team investigated how modifications to RNA impact the activity of SARS-CoV-2 RNA synthesis complex, also known as RNA dependent RNA polymerase (RdRp). Two types of RNA modifications, N1-methyladenosine (m1A) and N3-methylcytosine (m3C), interfere with the normal base-pairing in RNA. Surprisingly, m1A inhibits RdRp, while m3C can still be processed. These findings suggest that people whose epitranscriptomic systems can remove the methyl group from m1A in RNAs may be more susceptible to SARS-CoV-2.
Further research is needed to determine the impact of epitranscriptomics on viral infection symptoms. The data also provide insights into the development of new antiviral therapeutics.
- Effects of Epitranscriptomic RNA Modifications on the Catalytic Activity of SARS-CoV-2 Replication Complex,
Alexander Apostle, Yipeng Yin, Komal Chillar, Adikari M. D. N. Eriyagama, Reed Arneson, Emma Burke, Shiyue Fang, Yinan Yuan,
ChemBioChem 2023.
https://doi.org/10.1002/cbic.202300095
Also of Interest
- ChemistryViews Collection:
SARS-CoV-2 Virus – What We Know About Vaccines and Immunity